Source code of my master's thesis in context of Project Motiv. The thesis has two main contributions:
- Compression of atmospheric data using matrix approximation techniques
- Analysis of atmospheric data using clustering methods
- Create a virtual environment:
python -m venv myvenv - Activate virtual environment:
source myvenv/bin/activate - Install dependencies:
pip install -r requirements.txt - (Optional) Run unit tests:
python -m unittest discover -v -s ./test
For simple reconstruction of compressed data we provide iasi.Composition module.
The following minimal example demonstrates the reconstruction of water vapour averaging kernel:
from netCDF4 import Dataset
from iasi import Composition
# get required data
dataset = Dataset('/path/to/compressed/netcdf/file.nc')
number_of_levels = dataset['atm_nol'][...]
decomposed_group = dataset['state/WV/avk']
# use composition factory to reconstruct original data as masked array
composer = Composition.factory(decomposed_group)
wv_averaging_kernel = composer.reconstruct(number_of_levels)
print('Reconstructed Kernel shape: ', wv_averaging_kernel.shape)If you like to save the reconstructed variable into another Dataset, you can pass a target to the reconstruction method:
target_dataset = Dataset('reconstruction.nc', 'w')
composer.reconstruct(number_of_levels, target=target_dataset)
target_dataset.close()The reconstruction API automatically discovers the decomposition type (Singular Value Decomposition or Eigen Decomposition) by the group name. You may reconstruct matrices manually, but you should be careful: The reconstruction depends on decomposition type and dimensionality. The following snippet outlines the manual reconstruction of a single measurement of water vapour's averaging kernel.
from netCDF4 import Dataset
from iasi import Composition
# get required data
dataset = Dataset('/path/to/compressed/netcdf/file.nc')
# number of levels for first measurment
nol = dataset['atm_nol'][0]
# get decomposition components for first measurement
U = dataset['state/WV/avk/U'][0]
s = dataset['state/WV/avk/s'][0]
Vh = dataset['state/WV/avk/Vh'][0]
# reconstruct kernel matrix
rc = (U * s).dot(Vh)
# or in case of noise matrix
# rc = (Q * a).dot(Q.T)
print('Reconstructed kernel shape (single event):', rc.shape)
d = dataset.dimensions['atmospheric_grid_levels'].size
# reconstruct four matrices from single matrix
rc = np.array([
[rc[0:nol + 0, 0:nol + 0], rc[d:d + nol, 0:nol + 0]],
[rc[0:nol + 0, d:d + nol], rc[d:d + nol, d:d + nol]]
])
print('Reconstructed kernel after reshape (single event):', rc.shape)To avoid pitfalls we recommend using the iasi.Composition module!
This project manages processing of files with Luigi. Processing steps are implemented as Luigi Tasks. For task execution you need the luigi task scheduler, which can be started typing
luigid --logdir luigi-logs --background
The luigi scheduler backend should be available at http://localhost:8082/. You can stop luigi with killall luigid.
For testing purpose you can also pass --local-scheduler as a task parameter.
Luigi configuration settings are in luigi.cfg
For further details have a look at the Luigi Documentation.
Using command line interface
python -m luigi --module iasi DecompressDataset \
--file ./test/resources/MOTIV-single-event-compressed.nc \
--dst ./data \
[--local-scheduler] \
[--workers 1]
Using python module
import luigi
from iasi import DecompressDataset
task = DecompressDataset(file='test/resources/MOTIV-single-event-compressed.nc', dst='data')
luigi.build([task], local_scheduler=True, workers=1)Decompressing multiple files
import glob
import luigi
from iasi import DecompressDataset
files = glob.glob('src/of/files/*.nc')
tasks = [DecompressDataset(file=file, dst='data') for file in files]
luigi.build(tasks, local_scheduler=True, workers=1)If you pass the boolean parameter compress-upstream, the file specified with file is first compressed and then decompressed.
Using command line interface
python -m luigi --module iasi CompressDataset \
--file ./test/resources/MOTIV-single-event.nc \
--dst ./data \
[--local-scheduler] \
[--workers 1]
Using python module
import luigi
from iasi import CompressDataset
task = CompressDataset(file='test/resources/MOTIV-single-event.nc', dst='data')
luigi.build([task], local_scheduler=True, workers=1)For compression of multiple files have a look at slurm compression script. Warning: If you run many tasks on multiple nodes, the central luigi scheduler may be overloaded. As a workaround you can use the local scheduler. But in this case you have to take care for splitting date into chunks since there is no central component coordinating multiple compute node.
--file: file to process--dst: destination directory for task output--force: delete task output and execute again--force-upstream: delete all intermediate task output (excluding--file)--log-file: log task output to a file into destination directory (log file is automatically created)
Analysis of data requires conda setup (pip will not work):
- Create conda environment:
conda env create -f environment.yml - Activate conda environment:
conda activate iasi - (Optional) run analysis specific tests:
python -m unittest discover -v -s ./test -p 'analysis*.py'
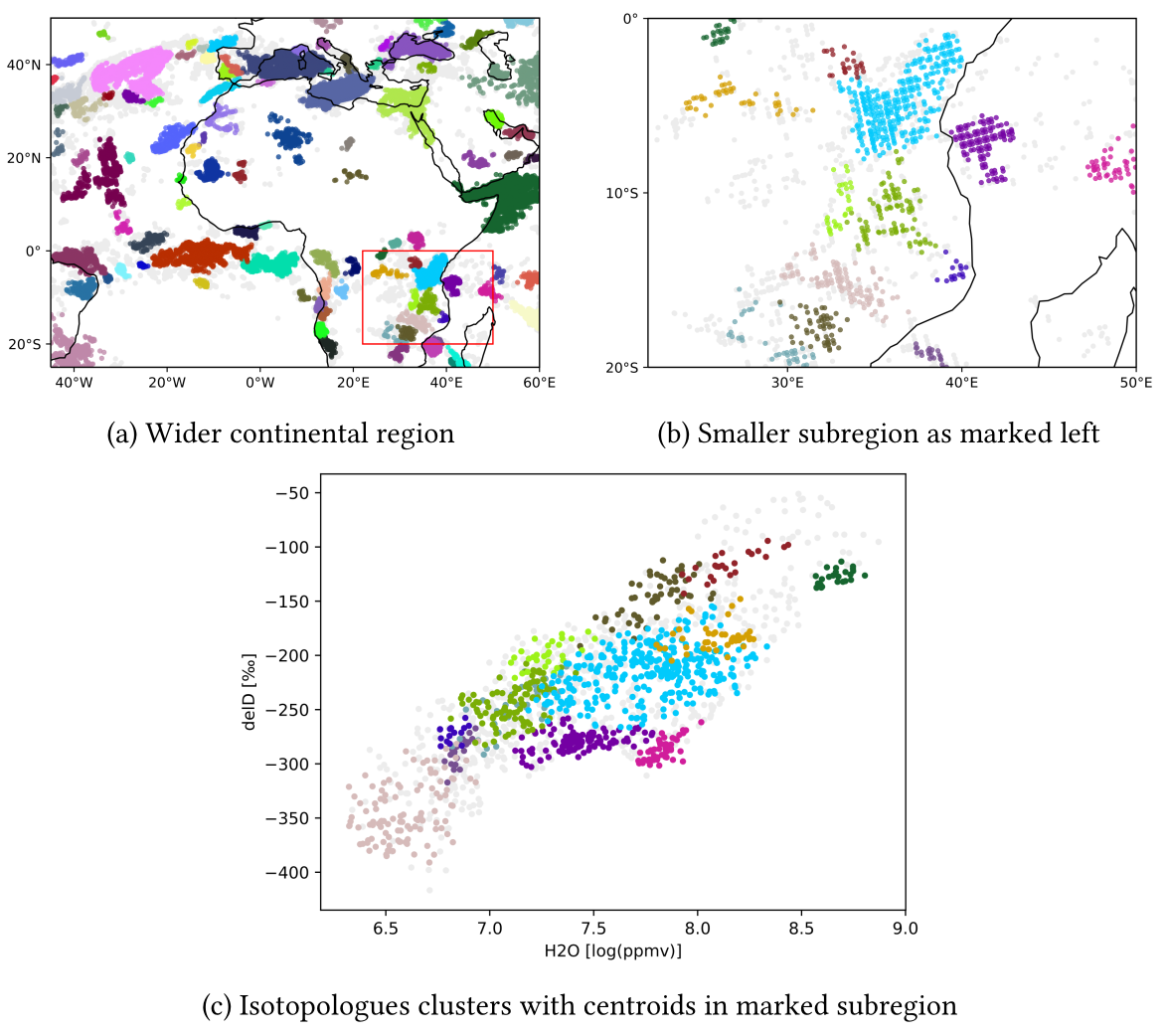
The pipeline.py file provides an example for spatio-temporal clustering.
With analysis.data.GeographicArea, you can specifiy the geographic extent of your analysis. By running the file
python pipeline.py
you receive something link this (depending on input data):
For quantitative analysis, you can perform GridSearch using grid_search.py:
python grid_search.py
Analogous to compression, analysis.aggregation.AggregateClusterStatistics is a luigi task that collects all clustered results
in a CSV including among others Davies-Bouldin Index , Silhoutte Index and DBCV (for HDBSCAN).
As shown in the example, you can pass custom parameters to the clustering algorithm as well as to the feature scaler.