GoPlantUML is an open-source tool developed to streamline the process of generating PlantUML diagrams from Go source code. With GoPlantUML, developers can effortlessly visualize the structure and relationships within their Go projects, aiding in code comprehension and documentation. By parsing Go source code and producing PlantUML diagrams, GoPlantUML empowers developers to create clear and concise visual representations of their codebase architecture, package dependencies, and function interactions. This tool simplifies the documentation process and enhances collaboration among team members by providing a visual overview of complex Go projects. GoPlantUML is actively maintained and welcomes contributions from the Go community.
Take a look at www.dumels.com. We have created dumels using this library.
Please, review the code of conduct here.
golang 1.17 or above
go get github.com/jfeliu007/goplantuml/parser
go install github.com/jfeliu007/goplantuml/cmd/goplantuml@latest
This will install the command goplantuml in your GOPATH bin folder.
goplantuml [-recursive] path/to/gofiles path/to/gofiles2
goplantuml [-recursive] path/to/gofiles path/to/gofiles2 > diagram_file_name.puml
Usage of goplantuml:
-aggregate-private-members
Show aggregations for private members. Ignored if -show-aggregations is not used.
-hide-connections
hides all connections in the diagram
-hide-fields
hides fields
-hide-methods
hides methods
-ignore string
comma separated list of folders to ignore
-notes string
Comma separated list of notes to be added to the diagram
-output string
output file path. If omitted, then this will default to standard output
-recursive
walk all directories recursively
-show-aggregations
renders public aggregations even when -hide-connections is used (do not render by default)
-show-aliases
Shows aliases even when -hide-connections is used
-show-compositions
Shows compositions even when -hide-connections is used
-show-connection-labels
Shows labels in the connections to identify the connections types (e.g. extends, implements, aggregates, alias of
-show-implementations
Shows implementations even when -hide-connections is used
-show-options-as-note
Show a note in the diagram with the none evident options ran with this CLI
-title string
Title of the generated diagram
-hide-private-members
Hides all private members (fields and methods)
goplantuml $GOPATH/src/github.com/jfeliu007/goplantuml/parser
// echoes
@startuml
namespace parser {
class Struct {
+ Functions []*Function
+ Fields []*Parameter
+ Type string
+ Composition []string
+ Extends []string
}
class LineStringBuilder {
+ WriteLineWithDepth(depth int, str string)
}
class ClassParser {
- structure <font color=blue>map</font>[string]<font color=blue>map</font>[string]*Struct
- currentPackageName string
- allInterfaces <font color=blue>map</font>[string]<font color=blue>struct</font>{}
- allStructs <font color=blue>map</font>[string]<font color=blue>struct</font>{}
- structImplementsInterface(st *Struct, inter *Struct)
- parsePackage(node ast.Node)
- parseFileDeclarations(node ast.Decl)
- addMethodToStruct(s *Struct, method *ast.Field)
- getFunction(f *ast.FuncType, name string)
- addFieldToStruct(s *Struct, field *ast.Field)
- addToComposition(s *Struct, fType string)
- addToExtends(s *Struct, fType string)
- getOrCreateStruct(name string)
- getStruct(structName string)
- getFieldType(exp ast.Expr, includePackageName bool)
+ Render()
}
class Parameter {
+ Name string
+ Type string
}
class Function {
+ Name string
+ Parameters []*Parameter
+ ReturnValues []string
}
}
strings.Builder *-- parser.LineStringBuilder
@enduml
goplantuml $GOPATH/src/github.com/jfeliu007/goplantuml/parser > ClassDiagram.puml
// Generates a file ClassDiagram.puml with the previous specifications
There are two different relationships considered in goplantuml:
- Interface implementation
- Type Composition
The following example contains interface implementations and composition. Notice how the signature of the functions
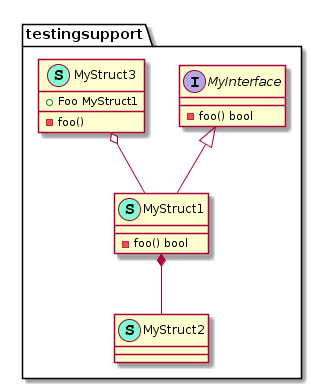
package testingsupport
//MyInterface only has one method, notice the signature return value
type MyInterface interface {
foo() bool
}
//MyStruct1 will implement the foo() bool function so it will have an "extends" association with MyInterface
type MyStruct1 struct {
}
func (s1 *MyStruct1) foo() bool {
return true
}
//MyStruct2 will be directly composed of MyStruct1 so it will have a composition relationship with it
type MyStruct2 struct {
MyStruct1
}
//MyStruct3 will have a foo() function but the return value is not a bool, so it will not have any relationship with MyInterface
type MyStruct3 struct {
Foo MyStruct1
}
func (s3 *MyStruct3) foo() {
}This will be generated from the previous code
@startuml
namespace testingsupport {
interface MyInterface {
- foo() bool
}
class MyStruct1 << (S,Aquamarine) >> {
- foo() bool
}
class MyStruct2 << (S,Aquamarine) >> {
}
class MyStruct3 << (S,Aquamarine) >> {
- foo()
+ Foo MyStruct1
}
}
testingsupport.MyStruct1 *-- testingsupport.MyStruct2
testingsupport.MyInterface <|-- testingsupport.MyStruct1
testingsupport.MyStruct3 o-- testingsupport.MyStruct1
@enduml
Diagram using www.dumels.com
For instructions on how to render these diagrams locally using plantuml please visit https://plantuml.com