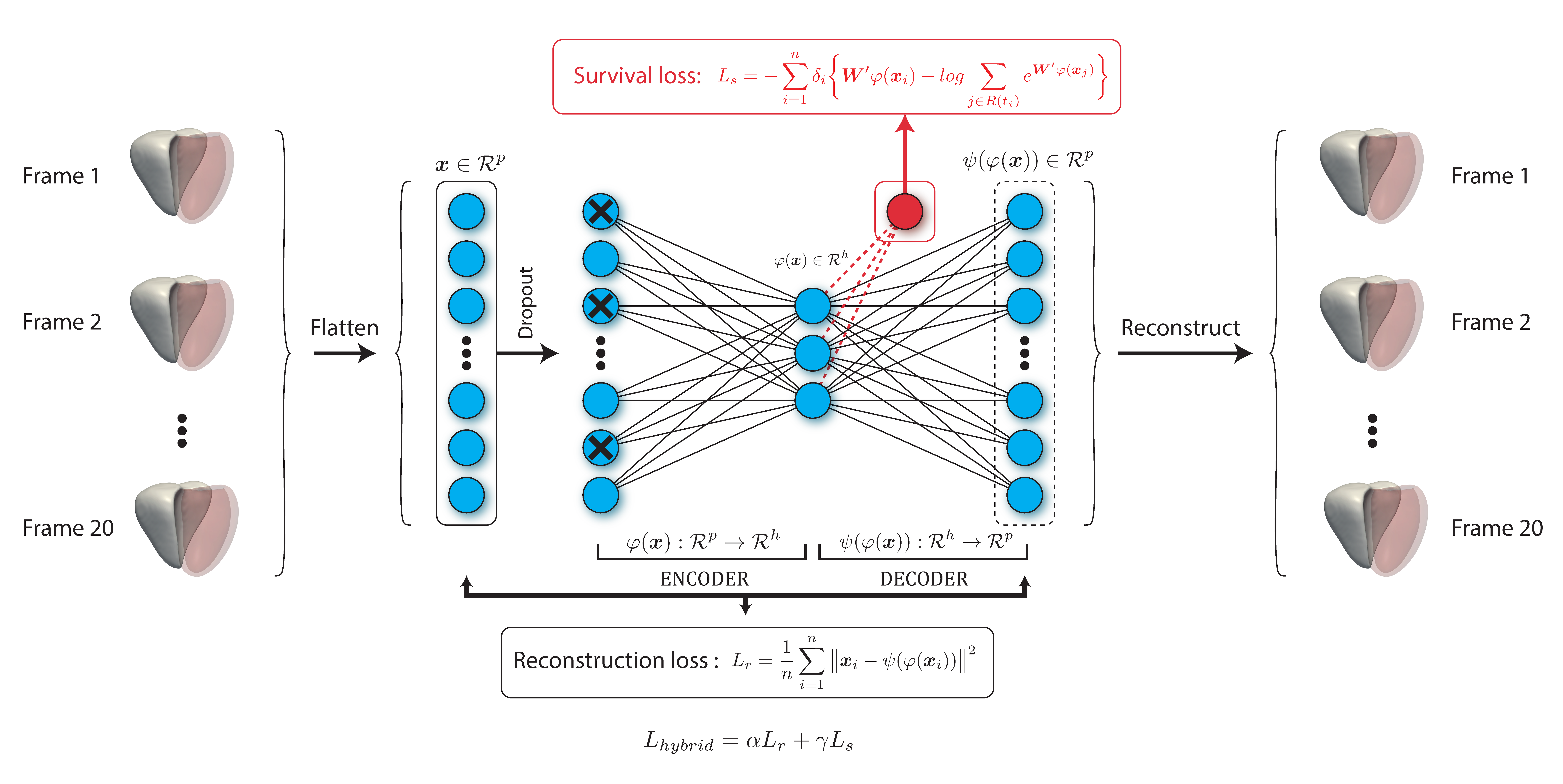
The code in this repository implements 4Dsurvival, a deep neural network for carrying out classification/prediction using 3D motion input data. The present implementation was trained using MRI-derived heart motion data and survival outcomes on pulmonary hypertension patients.
The files in this repository are organized into 3 directories:
- survival4D : contains base functions for fitting the 2 types of statistical models used in our paper: 4Dsurvival (supervised denoising autoencoder for survival outcomes) and a penalized Cox Proportional Hazards regression model.
- demo
- demo/scripts: contains functions for the statistical analyses carried out in our paper:
- Training of DL model - demo/scripts/demo_hypersearch_nn.py
- Default configuration used to train DL model - demo/scripts/default_nn.conf
- Default configuration used to cox reg model - demo/scripts/default_cox.conf
- Generation of Kaplan-Meier plots - demo/scripts/demo_KMplot.py
- statistical comparison of model performance - demo/scripts/demo_modelcomp_pvalue.py
- Bootstrap internal validation - demo/scripts/demo_validate.py
- demo/notebooks: contains ipython notebooks for demoing the above scripts.
- demo/scripts: contains functions for the statistical analyses carried out in our paper:
- data : contains simulated data on which functions from the
demodirectory can be run.
To run the code in the demo directory, we provide a Binder interface (for the Jupyter notebooks) and a Docker container (for the corresponding Python scripts). Below are usage instructions:
The Jupyter notebooks in the demo directory are hosted on Binder, which provides an interactive user interface for executing Jupyter notebooks. Click the link provided below for access:
You can also edit code, change parameters, and re-run the analysis interactively in Code Ocean.
A Docker image is available for running the code available in the demo directory. This image contains a base Ubuntu Linux operating system image set up with all the libraries required to run the code (e.g. Tensorflow, Keras, Optunity, etc.). The image contains all the code, as well as simulated cardiac data on which the code can be run.
Running our 4Dsurvival Docker image requires installation of the Docker software, instructions are available at https://docs.docker.com/install/
Once the Docker software has been installed, our 4Dsurvival Docker image can be pulled from the Docker hub using the following command:
docker pull lisurui6/4dsurvival-gpu:1.1
Once the image download is complete, open up a command-line terminal. On Windows operating systems, this would be the Command Prompt (cmd.exe), accessible by opening the Run Command utility using the shortcut key Win+R and then typing cmd. On Mac OS, the terminal is accessible via (Finder > Applications > Utilities > Terminal). On Linux systems, any terminal can be used.
Once a terminal is open, running the following command:
docker images
should show lisurui6/4dsurvival-gpu:1.0 on the list of Docker images on your local system
To be able to utilise GPU in docker container, run
nvidia-docker run -it lisurui6/4dsurvival:1.1
This launches an interactive linux shell terminal that gives users access to the image's internal file system. This file system contains all the code in this repository, along with the simulated data on which the code can be run. Typing
ls -l
will list all the folders in the working directory of the Docker image (/4DSurv). You should see the 3 main folders code, data and demo, which contain the same files as the corresponding folders with the same name in this github repository.
Dockerfile for build the GPU image is described in Dockerfile.
In the docker image, survival4D has already installed, so that you can run the following python command anywhere.
If you are running outside of docker, and want to install the package, from the 4Dsurvival directory, do:
python setup.py develop
develop command allows you to makes changes to the code and do not need to reinstall for the changes to be applied.
This section is dedicated on instructions and best practices to run 4DS experiments with docker. First, although the docker image contains 4DS code, it is best to mount the code from your local file system, so that you can easily change the code using your favorite editor, and push/pull changes to/from github.
Second, to have access to your data inside the docker image, you need to mount your data from your local file system as well.
Lastly, you need to mount a experiment directory, where your experiment .conf file is and your experiment output will be.
A typical command for launching a docker container for running experiment would be
nvidia-docker run -it -v /path-to-your-data:/data -v /path-to-your-4ds-code:/4DSurvival -v /path-to-your-experiment-dir:/exp-dir/ lisurui6/4dsurvival:1.1
Now outside the docker container, you need to create a .conf file in /path-to-your-experiment-dir/exp-name.conf, similar to demo/scripts/default_nn.conf or
demo/scripts/default_cox.conf. Change the data_path and output_dir to
experiment {
data_path = "/data/data-filename.pkl"
output_dir = "/exp-dir/exp-name/"
Then inside the docker container,
cd /4DSurvival
python setup.py develop
cd demo/scripts/
CUDA_VISIBLE_DEVICES=0 python demo_validate_nn.py -c /exp-dir/exp-name.conf
.conf file specifies experiment parameters, including experiment configuration, hyperparam search range, and model arguments (for neural network only).
Experiment configurations are under the umbrella of experiment group. It includes data path, output directory, hyperparam search method, etc.
Hyperparam search ranges are under the umbrella of hypersearch group. It indicates param names and their ranges.
Model arguments (for nn only), are parameters of the model that are fixed. Either they cannot be searched on, or we do no wish to search them.
Between hypersearch and model, they should contains all the arguments needed for constructing a model.
Below we will demonstrate how to perform (within the Docker image) the following analyses:
- Train deep learning network
- Train and validate conventional parameter model
From the 4Dsurvival directory, navigate to the demo/scripts directory by typing:
cd demo/scripts
ls -l
The demo_hypersearch_nn.py file should be visible. This executes a hyperparameter search (see Methods section in paper)
for training of the 4Dsurvival deep learning network. A demo of this code (which uses simulated input data) can now
be run (WARNING: on most machines, this will take several hours to complete):
python3 demo_hypersearch_nn.py
Also under the demo/scripts folder, the demo_validate_nn.py file should be visible. This executes the bootstrap-based
approach for training and internal validation of the deep learning network. This can be run (WARNING: this may take days to complete):
python3 demo_validate_nn.py
demo_validate_nn.py will output the conf that it uses, all the hyperparams searched and found, and figures of c-index
vs more bootstrap samples in a output_dir, which by default is output folder under the data path directory.
Both demo_hypersearch_nn.py and demo_validate_nn.py takes a .conf file as an input argument.
If not provided, by default, it will uses demo/scripts/default_nn.conf.
Such .conf file has the following structure:
experiment {
data_path = "../../data/inputdata_DL.pkl"
output_dir = None # If it is None, output dir would be `output` folder under the data path directory
batch_size = 16
n_epochs = 100
n_evals = 50
n_bootstraps = 100
n_folds = 6
model_name = "baseline_autoencoder" # must be one of the function names in model_factory function of survival4D/models
}
# hyperparams search range define here.
# Notice that value name has to match input arguments' names, defined in survival4D/models
hypersearch {
lr_exp = [-6., -4.5]
loss_alpha = [0.3, 0.7]
dropout = [0.1, 0.5]
num_ae_units1 = [75, 250]
num_ae_units2 = [5, 20]
l1_reg_lambda_exp = [-7, -4]
}
If a user wants to change any value in the .conf file, such as data_path for hypersearch. Create a user.conf file, and
copy from default_nn.conf, before making change to the values. Afterwards, run demo_validate_nn.py or demo_hypersearch_nn.py:
python3 demo_validate_nn.py -c /path-to-user.conf
python3 demo_hypersearch_nn.py -c /path-to-user.conf
Under the demo/scripts folder, the demo_validate.py file should be visible. This executes the bootstrap-based
approach for training and internal validation of the Cox Proportional Hazards model for conventional (volumetric)
parameters. A demo of this code (which uses simulated input data) can now be run :
python3 demo_validate.py
It takes a .conf file as an input argument. If not provided, by default, it will uses demo/scripts/default_cox.conf.
Such .conf file has the following structure:
experiment {
data_path = "../../data/inputdata_conv.pkl"
output_dir = None # If it is None, output dir would be `output` folder under the data path directory
batch_size = 16
n_epochs = 100
n_evals = 50
n_bootstraps = 100
n_folds = 6
model_name = "cox" # must be one of the function names in model_factory function of survival4D/models
}
# hyperparams search range define here.
# Notice that value name has to match input arguments' names, defined in survival4D/models
hypersearch {
penalty_exp = [-2, 1]
}
If a user wants to change any value in the .conf file, such as data_path for hypersearch. Create a user.conf file, and
copy from default_nn.conf, before making change to the values. Afterwards, run demo_validate.py:
python3 demo_validate.py -c /path-to-user.conf
Bello GA, Dawes TJW, Duan J, Biffi C, de Marvao A, Howard LSGE, Gibbs JSR, Wilkins MR, Cook SA, Rueckert D, O'Regan DP. Deep-learning cardiac motion analysis for human survival prediction. Nature Machine Intelligence 1, 95–104 (2019).
Duan J, Bello G, Schlemper J, Bai W, Dawes TJ, Biffi C, de Marvao A, Doumou G, O’Regan DP, Rueckert D. Automatic 3D bi-ventricular segmentation of cardiac images by a shape-refined multi-task deep learning approach. IEEE Transactions on Medical Imaging 38(9), 2151-2164 (2019).