Author: Yixue Feng
- Create conda environment using
conda env create -f environment.yml. - Run BundleCleaner with the default parameters,
python src/BundleCleanerV2.py -i test_bundles/AF_L.trk -o test_bundles/AF_L_proc.trk. Add-vflag for verbose output. - Python implementation of select bundle shape metrics defined in DSI Studio [1] are available at
src/BundleInfo.py(original implementation). - Sample bundle from processed PPMI data for testing is provided at
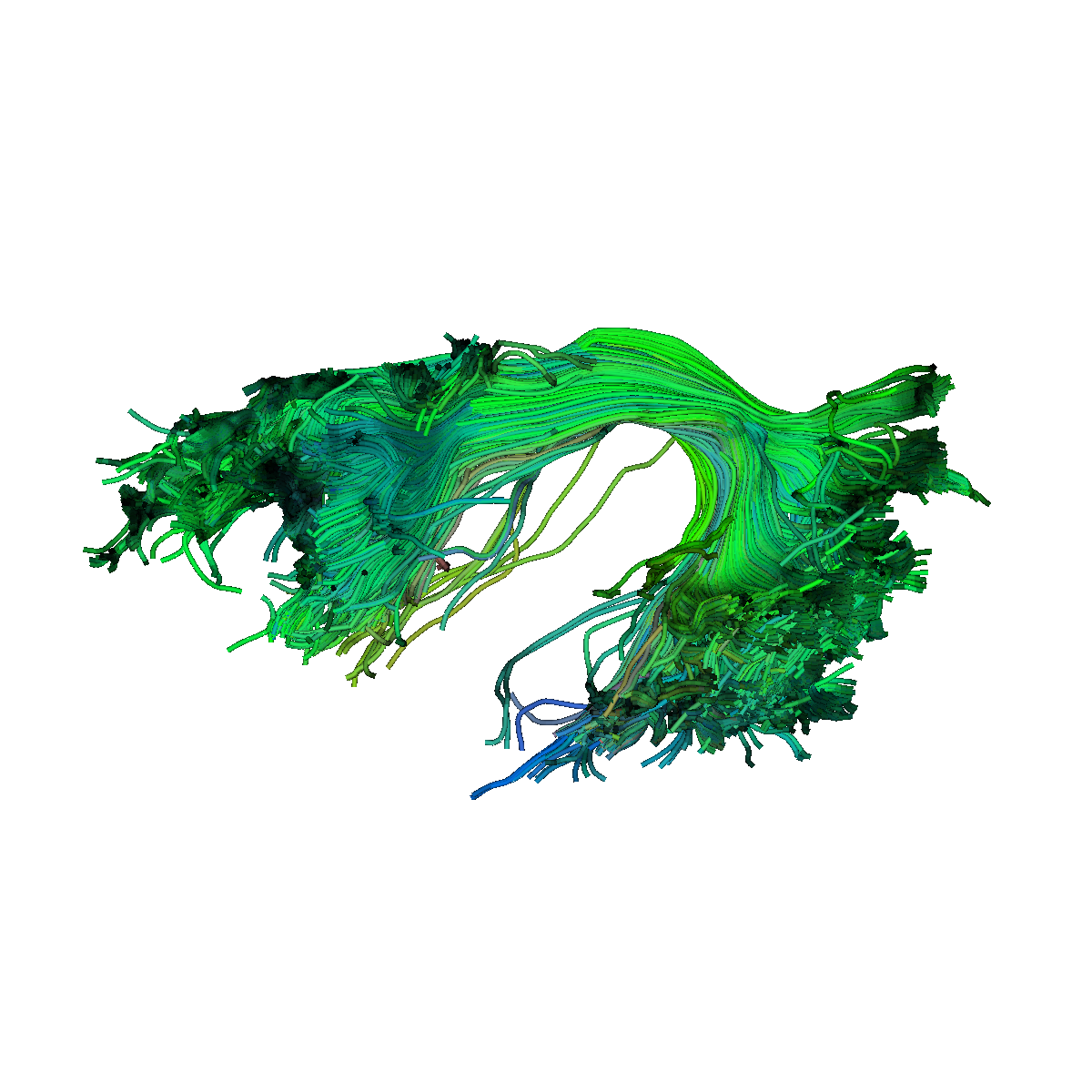
test_bundles/AF_L.trk.
This work was accepted to the MICCAI 2023 CDMRI workshop. Preprint available at https://www.biorxiv.org/content/10.1101/2023.08.19.553990v1.
@misc{feng_bundlecleaner_2023,
title = {{BundleCleaner}: {Unsupervised} {Denoising} and {Subsampling} of {Diffusion} {MRI}-{Derived} {Tractography} {Data}},
copyright = {All rights reserved},
shorttitle = {{BundleCleaner}},
url = {https://www.biorxiv.org/content/10.1101/2023.08.19.553990v1},
doi = {10.1101/2023.08.19.553990},
language = {en},
urldate = {2023-08-21},
publisher = {bioRxiv},
author = {Feng, Yixue and Chandio, Bramsh Q. and Villalon-Reina, Julio E. and Thomopoulos, Sophia I. and Joshi, Himanshu and Nair, Gauthami and Joshi, Anand A. and Venkatasubramanian, Ganesan and John, John P. and Thompson, Paul M.},
month = aug,
year = {2023},
note = {Accepted to MICCAI 2023 CDMRI Workshop.}
}
[1] F.-C. Yeh, “Shape analysis of the human association pathways,” NeuroImage, vol. 223, p. 117329, Dec. 2020, doi: 10.1016/j.neuroimage.2020.117329.
[2] B. Q. Chandio et al., “Bundle analytics, a computational framework for investigating the shapes and profiles of brain pathways across populations,” Sci Rep, vol. 10, no. 1, p. 17149, Dec. 2020, doi: 10.1038/s41598-020-74054-4.