This repository contains the source code for the ICCV 2023 paper — Robust e-NeRF: NeRF from Sparse & Noisy Events under Non-Uniform Motion, which is mainly built on the PyTorch Lightning framework. Robust e-NeRF is a novel method to directly and robustly reconstruct NeRFs from moving event cameras under various real-world conditions, especially from sparse and noisy events generated under non-uniform motion.
If you find Robust e-NeRF useful for your work, please consider citing:
@inproceedings{low2023_robust-e-nerf,
title = {Robust e-NeRF: NeRF from Sparse & Noisy Events under Non-Uniform Motion},
author = {Low, Weng Fei and Lee, Gim Hee},
booktitle = {Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)},
year = {2023}
}We recommend using Conda to set up an environment with the appropriate dependencies for running Robust e-NeRF, as follows:
git clone https://github.com/wengflow/robust-e-nerf.git
cd robust-e-nerf
conda env create -f environment.ymlIf a manual installation is preferred, the list of dependencies can be found in environment.yml.
Our synthetic experiments are performed on a set of sequences simulated using an improved ESIM event camera simulator with different camera configurations on NeRF Realistic Synthetic
To run Robust e-NeRF on our synthetic dataset:
- Setup the dataset according to the official instructions
- Preprocess each sequence in the raw dataset with:
python scripts/preprocess_esim.py <sequence_path>/esim.conf <sequence_path>/esim.bag <sequence_path>
Our qualitative real experiments are performed on the mocap-1d-trans, mocap-desk2 and office_maze sequences of the TUM-VIE dataset, which are setup as follows:
- Download the following raw data for each sequence and calibration files into a common folder:
<sequence_name>-events_left.h5<sequence_name>-vi_gt_data.tar.gzcamera-calibration{A, B}.jsonmocap-imu-calibration{A, B}.json
- Uncompress all
<sequence_name>-vi_gt_data.tar.gzand then remove them - Preprocess each sequence in the raw dataset with:
Note that we trim the end of the
python scripts/tum_vie_to_esim.py <sequence_name> <raw_dataset_path> <preprocessed_dataset_path>
offize-mazesequence with the additional argument of--end_timestamp 20778222508
Train, validate or test Robust e-NeRF with:
python scripts/run.py {train, val, test} <config_file_path>In the configs/ folder, we provide two sets of configuration files:
{train, test}/synthetic.yaml{train, test}/{mocap-1d-trans, mocap-desk2, office_maze}.yaml
used to train or test Robust e-NeRF for the synthetic and real experiments, respectively.
The specific experimental setting described in the configuration files are given as follows:
| Configuration File | Experiment | Sequence | Opt. |
Opt. |
|
|---|---|---|---|---|---|
synthetic.yaml |
Synthetic |
ficus under the easy/default setting |
✗ | ✗ | ✗ |
<sequence_name>.yaml |
Real | <sequence_name> |
✓ | ✓ | ✓ |
You should modify the following parameters in the given configuration files to reflect the correct or preferred paths:
data.dataset_directory: Path to the preprocessed sequencemodel.checkpoint_filepath: Path to the pretrained modellogger.save_dir: Preferred path to the logs
To reproduce our synthetic experiment results under any specific setting, as reported in the paper, you should modify {train, test}/synthetic.yaml as follows:
| Experimental Setting | Parameter(s) To Be Modified |
|---|---|
| Sequence | data.dataset_directory |
| Opt. |
model.contrast_threshold.freeze |
| Opt. |
model.refractory_period.freeze |
| w/ or w/o |
loss.weight.log_intensity_grad |
You should also modify model.checkpoint_filepath and logger.name accordingly.
Note that in our synthetic experiments, when the contrast threshold
Please refer to the Robust e-NeRF paper for more details.




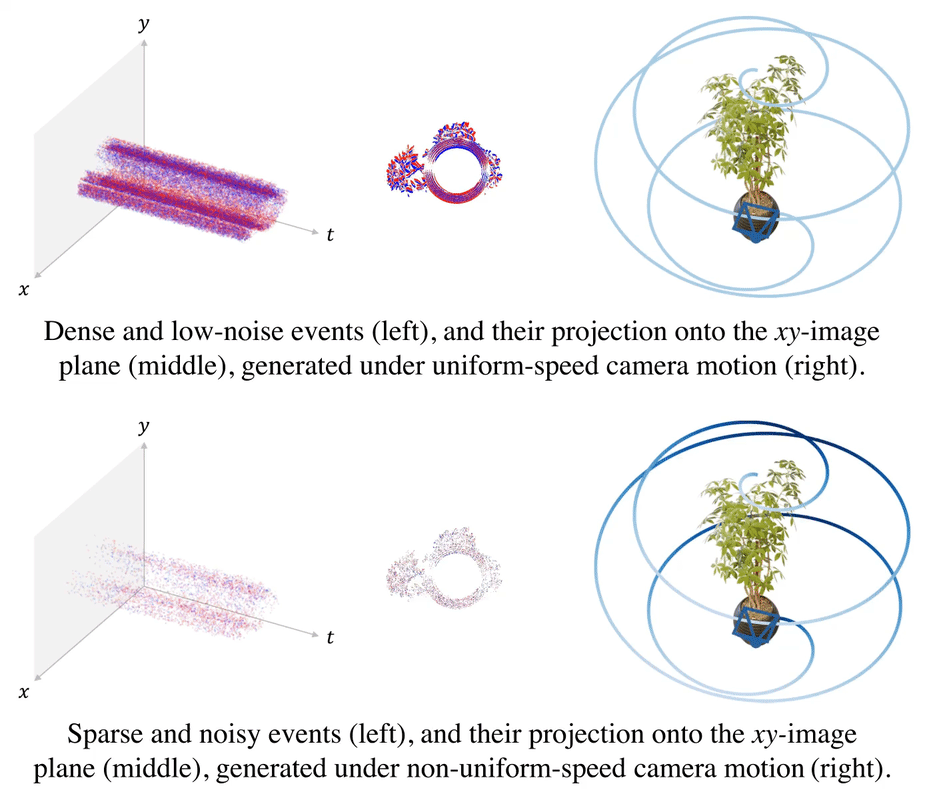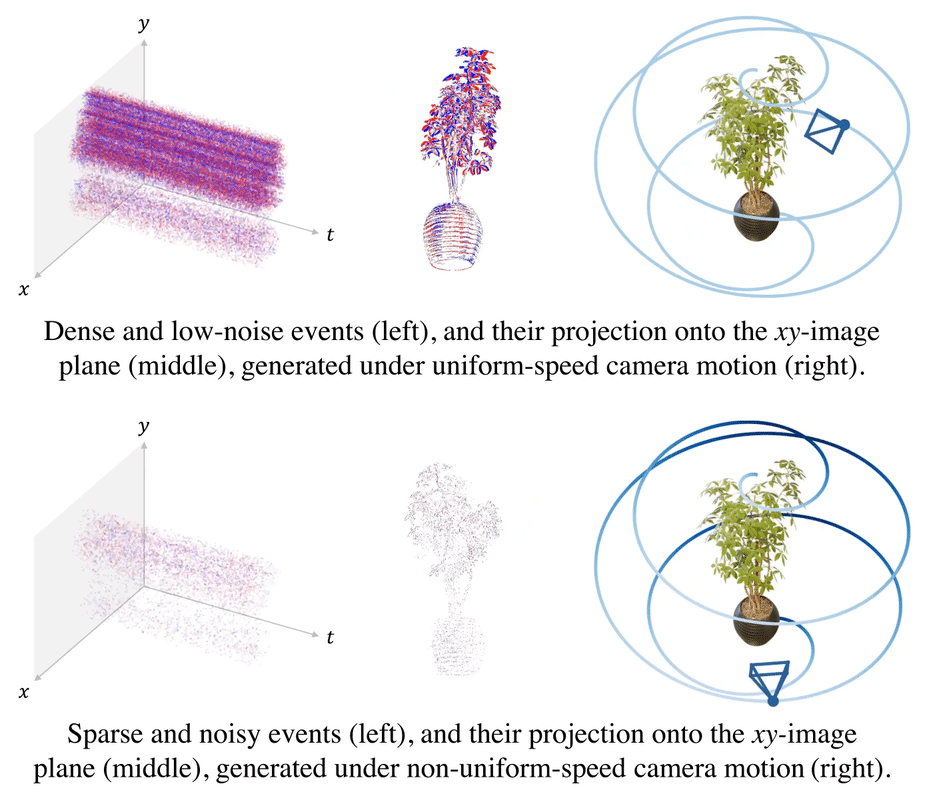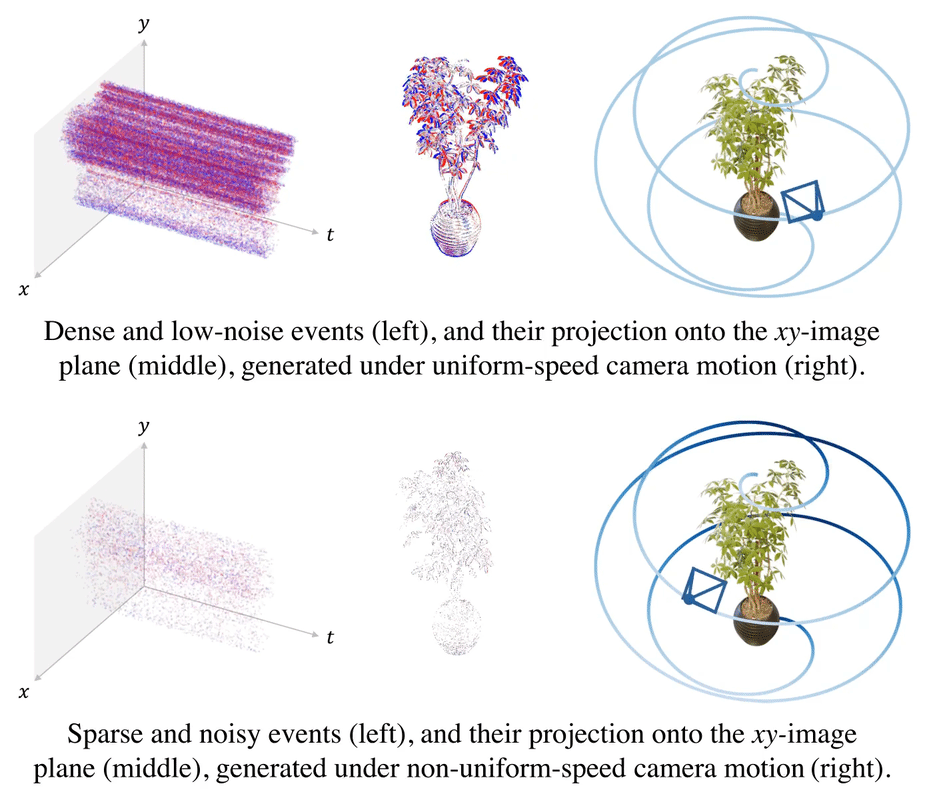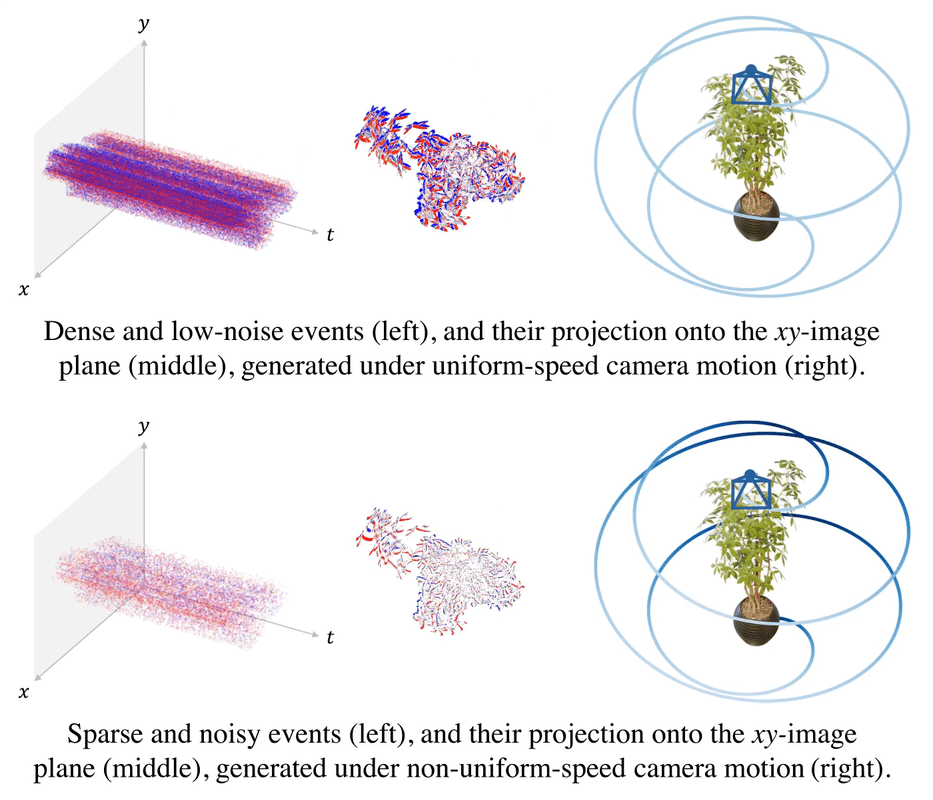
.gif)