- 3D Segmentation & Classification with Keras
- Fine preprocessing with scikit-image
- Fine visualization for clarification
- Modified UNet for segmentation
- Modified VGG/Inception/ResNet/DenseNet for classification ensemble
- Fine hyperparameter tunning with both models and training process.
- config.py # good practice to centralize hyper parameters
- preprocess.py # Step 1, preprocess, store numpy/meta 'cache' at ./preprocess/
- train_segmentation.py # Step 2, segmentation with UNet Model
- model_UNet.py # UNet model definition
- train_classificaion.py # Step 3, classificaiton with VGG/Inception/ResNet/DenseNet
- model_VGG.py # VGG model definition
- model_Inception.py # Inception model definition
- model_ResNet.py # ResNet model definition
- model_DenseNet.py # DenseNet model definition
- generators.py # generator for segmentation & classificaiton models
- visual_utils.py # 3D visual tools
- dataset/ # dataset, changed in config.py
- preprocess/ # 'cache' preprocessed numpy/meta data, changed in config.py
- train_ipynbs # training process notebooks
- use
SimpleITKto read CT files, process, and store into cache with numpy arrays - process with
scikit-imagelib, try lots of parameters for best cutting- binarized
- clear-board
- label
- regions
- closing
- dilation
- collect all meta information(seriesuid, shape, file_path, origin, spacing, coordinates, cover_ratio, etc.) and store in ONE cache file for fast training init.
- see preprocessing in
/train_ipynbs/preprocess.ipynbfile
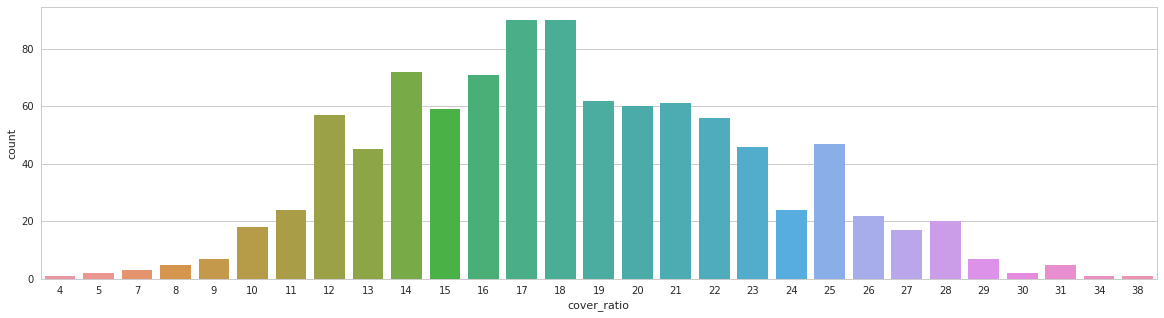
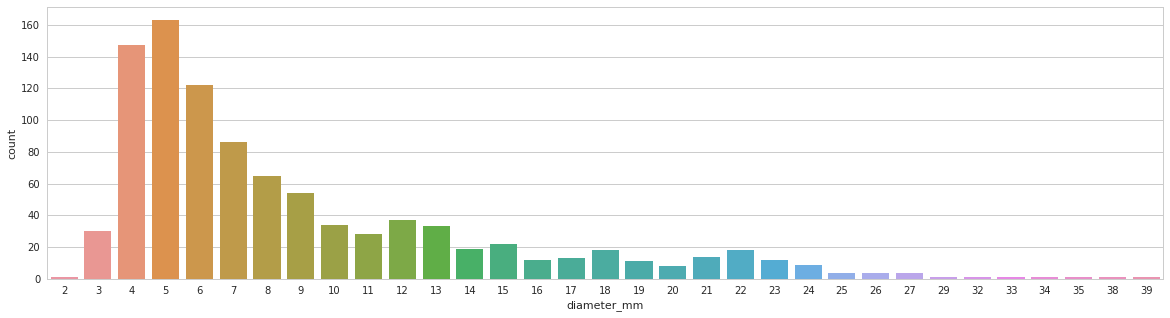
Distribution of the lung part takes on a whole CT.
Tumor size distribution
- A simplified and full UNet both tested.
dice_coef_lossas loss function.- Periodically evaluate model with lots of metrics, which helps a lot to understand the model.
- 30% of negative sample, which has no tumor, for generalization.
- Due to memory limitation, 16 batch size used.
- A simplified and full VGG model both tested. Use simplified VGG as baseline.
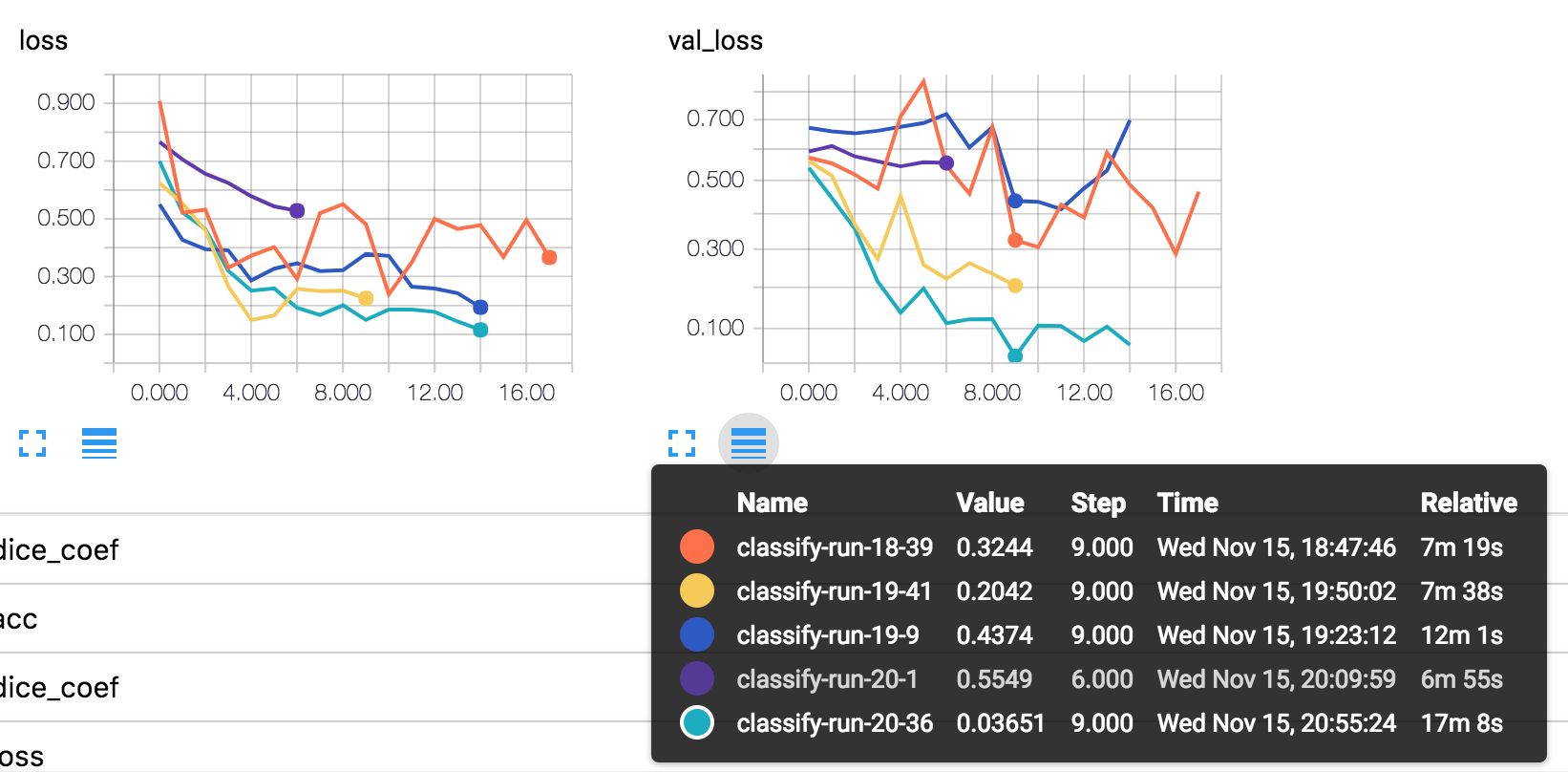
Pictures tells that: hyperparameter tunning really matters.
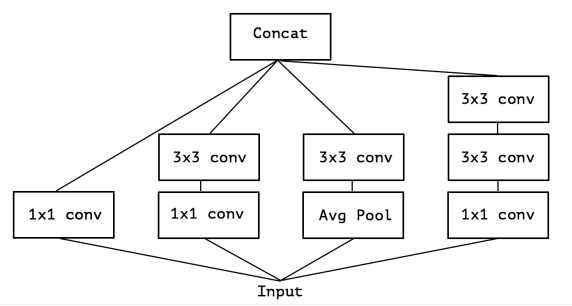
- A simplified Inception-module based network, with each block has 4-5 different type of conv.
- 1*1*1 depth-size seperable conv
- 1*1*1 depth-size seperable conv, then 3*3*3 conv_bn_relu
- 1*1*1 depth-size seperable conv, then 2 3*3*3 conv_bn_relu
- AveragePooling3D, then 1*1*1 depth-size seperable conv
- (optional in config) 1*1*1 depth-size seperable conv, and (5, 1, 1), (1, 5, 1), (1, 1, 5) spatial separable convolution
- Concatenate above.
- use
bottleneckblock instead ofbasic_blockfor implementation. - A
bottleneckresidual block consists of:- (1, 1, 1) conv_bn_relu
- (3, 3, 3) conv_bn_relu
- (1, 1, 1) conv_bn_relu
- (optional in config) kernel_size=(3, 3, 3), strides=(2, 2, 2) conv_bn_relu for compression.
- Add(not Concatenate) with input
- Leave
RESNET_BLOCKSas config to tune
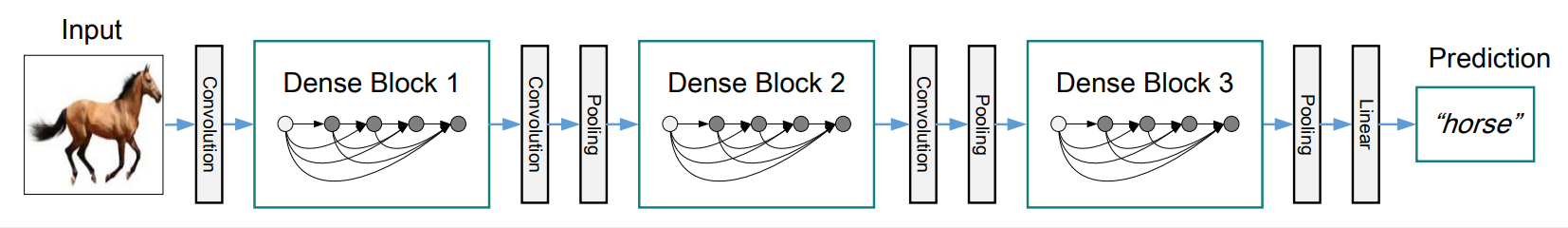
DenseNetdraws tons of experience from origin paper. https://arxiv.org/abs/1608.06993- 3 dense_block with 5 bn_relu_conv layers according to paper.
- transition_block after every dense_block, expcet the last one.
- Optional config for DenseNet-BC(paper called it): 1*1*1 depth-size seperable conv, and transition_block compression.
- Learning rate:
3e-5works well for UNet,1e-4works well for classification models. - Due to memory limitation, 16 batch size used.
- Data Augumentation: shift, rotate, etc.
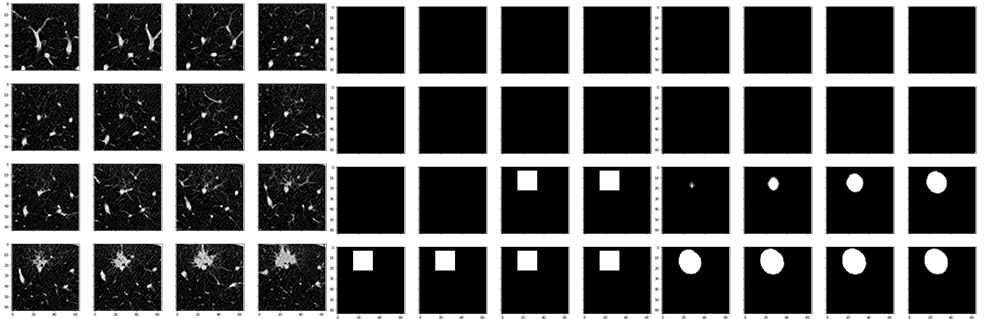
- Visualization cannot be more important!!!
- coord(x, y, z) accord to (width, height, depth), naughty bugs.
- Put all config in one file save tons of time. Make everything clean and tidy
- Disk read is bottle neck. Read from SSD.
- Different runs has different running log dirs, for better TensorBoard visualization. Make it like
/train_logs/<model-name>-run-<hour>-<minute>. - Lots of debug options in config file.
- 4 times probability strengthened for tumors < 10mm, 3 for tumor > 10mm and < 30mm, keep for > 30mm. Give more focus on small tumors, like below.