Started on Aug 20, 2019
Tranfer function analysis of blood pressure and CBFv for studying autoregulation in cerebral arteries.
Here is the BOX folder with all the code, as well as a cohort of subjects already done which give an idea of how things should be set up: https://upenn.app.box.com/folder/84579221507
Convert the xlsx files into tab-separated-value files
- Assume the excel files are named like so:
RECAST H026 RIGHT ACA.xlsx
-
Put the excel files into one folder. Put convert.sh and convert_excel_to_tsv.py in the same folder.
-
In the terminal, run
chmod +x convert.shto make the script executable -
Make sure you have Python 3 and pandas installed. If not, do pip install pandas from the command line
-
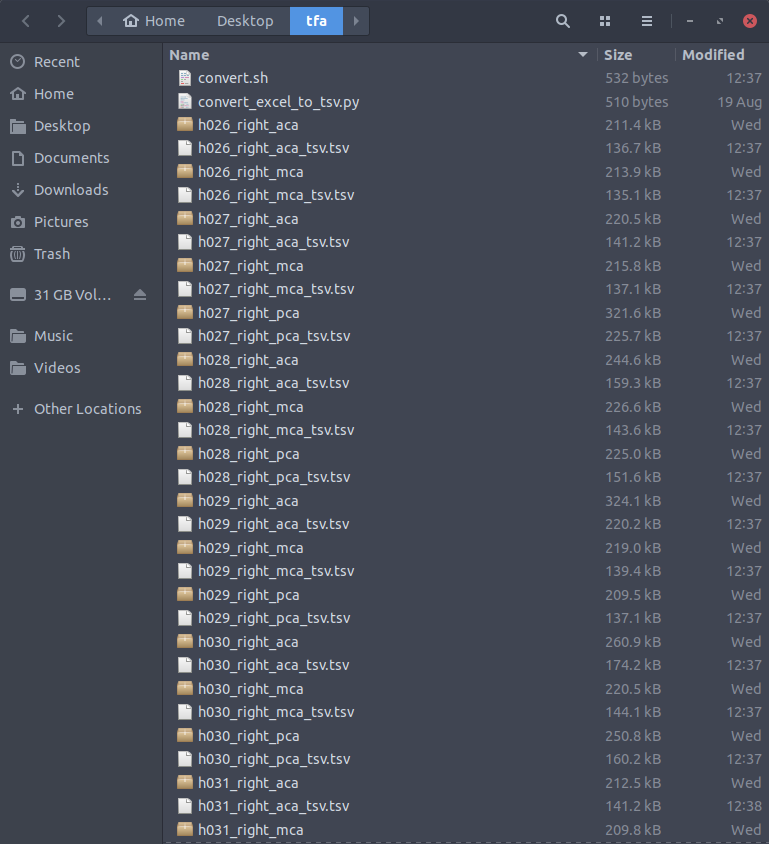
Run
./convert.sh. This will convert all the excel files into tsvs and rename them so that there are underscores in between subject name, hemisphere, and artery, resulting in a directory that looks like this:
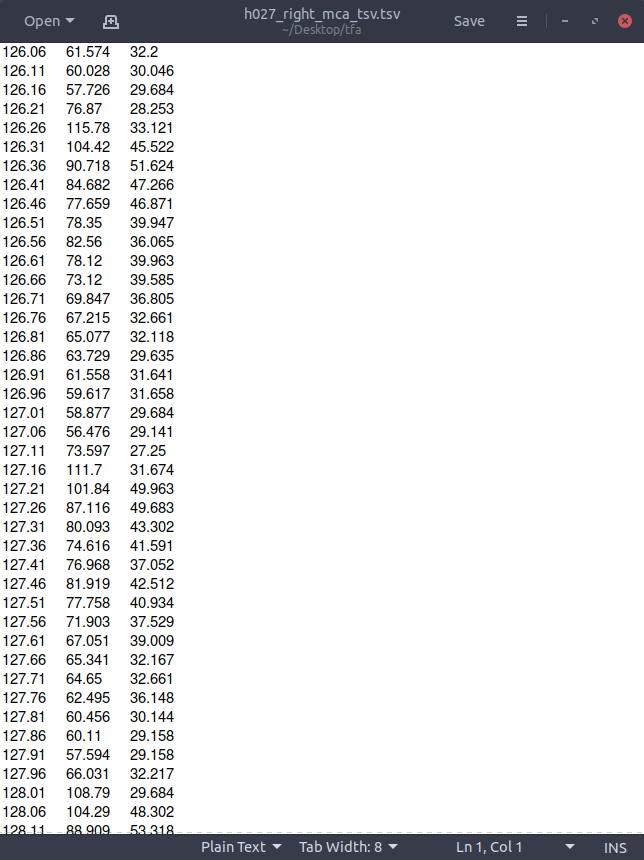
With tsv files that look like this:
Prep the transfer function analysis.
-
If not done already, make a folder with these sub-folders and files
-
Copy all the tsv files into Inputs
-
In outputs, make a folder with each subject name (e.g. h019) by hand or run makedirs.sh in the directory and change the numbers at the top of that script to reflect the subjects you want.
-
The matlab file, run_tfa.m, performs the analysis by parsing the name, hemisphere and artery for a given tsv file, getting the time, ABP, and CBF time series from the columns in the file and sending that data to the tfa_car.m program which does the heavy lifting of the transfer function analysis. It then filters out the fields we want and arranges them into key-value pairs which it saves as a text file. It also saves the figures as .figs (which are only openable in matlab...we’ll convert them later to PNG)
-
IMPORTANT: in run_tfa.m, there are paths defined on line 4, line 74 and line 86. Edit these to your own path
-
Since run_tfa.m only processes one subject at a time, I wrote a wrapper script to loop through all the subjects and call Matlab to run run_tfa.m for each one. This is batch_run_tfa.sh. NOTE that it will loop through ALL subjects in the input folder. If some of them are already done, the simplest solution would be to temporarily put them in a different folder, run batch_run_tfa.sh, and then put them back in the original inputs folder.
-
IMPORTANT: like above, edit the paths at the beginning of batch_run_tfa.sh to reflect your setup.
-
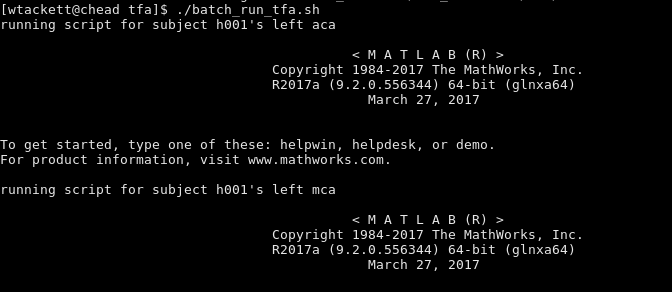
Run ./batch_run_tfa.sh. You’ll start to see something like this:
-
All the results will be saved in each subject’s folder in outputs/
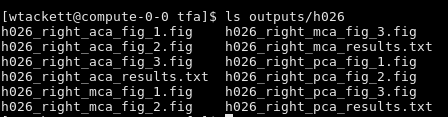
-
There will be a text file for each artery and side that looks like this:
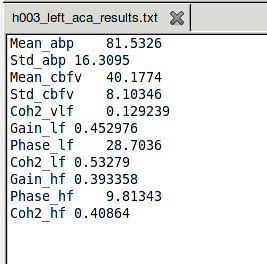
-
And there will be three figures.
-
Since they’re .fig files and completely useless, we convert them to the useful PNG format, using batch_convert.sh, which runs export_figs.m to do the conversion for each figure.
-
IMPORTANT: like above, edit the paths at the beginning of batch_convert.sh to reflect your setup.
-
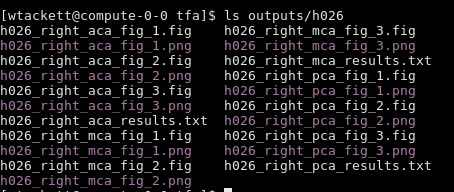
Now the output/h0xx folder looks like this:
- And we’re done! You can load those text files into an excel file and they should fit to the cells automatically and they’re ready to do group-level stats on.