PyTorch implementation of Noisy Natural Gradient as Variational Inference.
- Python 3
- Pytorch
- visdom
- This paper is about how to optimize bayesian neural network which has matrix variate gaussian distribution.
- This implementation contains Noisy Adam optimizer which is for Fully Factorized Gaussian(FFG) distribution, and Noisy KFAC optimizer which is for Matrix Variate Gaussian(MVG) distribution.
- These optimizers only work with bayesian network which has specific structure that I will mention below.
- Currently only linear layer is available.
- I addded a lr scheduler to noisy KFAC because loss is exploded during training. I guess this happens because of slight approximation.
- For MNIST training noisy KFAC is 15-20x slower than noisy Adam, as mentioned in paper.
- I guess the noisy KFAC needs more epochs to train simple neural network structure like 2 linear layers.
Currently only MNIST dataset are currently supported, and only fully connected layer is implemented.
model: Fully Factorized Gaussian(FFG) or Matrix Variate Gaussian(MVG)n: total train dataset size. need this value for optimizer.eps: parameter for optimizer. Default to 1e-8.initial_size: initial input tensor size. Default to 784, size of MNIST data.label_size: label size. Default to 10, size of MNIST label.
More details in option_parser.py
$ python train.py --model=FFG --batch_size=100 --lr=1e-3 --dataset=MNIST
$ python train.py --model=MVG --batch_size=100 --lr=1e-2 --dataset=MNIST --n=60000
- To visualize intermediate results and loss plots, run
python -m visdom.serverand go to the URL http://localhost:8097
$ python test.py --epoch=20
- network is consist of 2 linear layers.
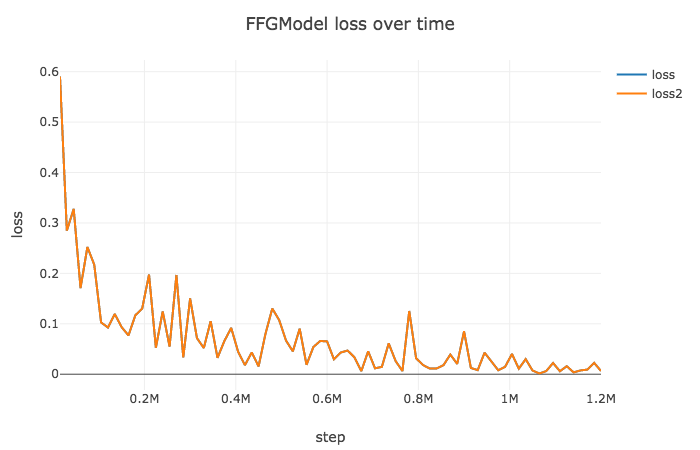
- FFG optimized by noisy Adam :
epoch20,lr1e-3
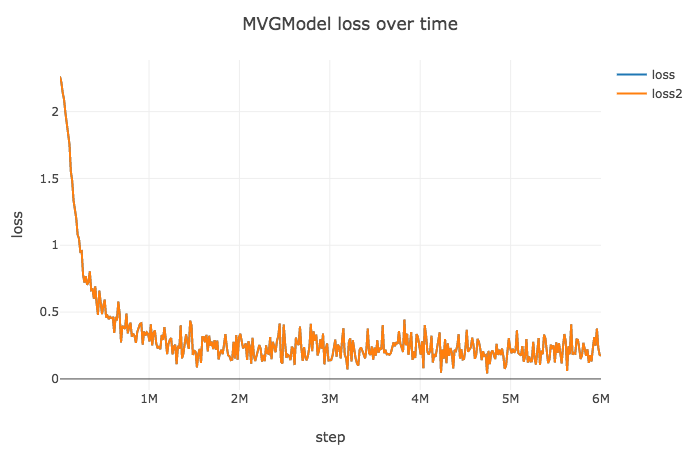
- MVG optimized by noisy KFAC :
epoch100,lr1e-2, decay 0.1 for every 30 epochs - Need to tune learning rate.
- Optimizing parameter procedure is consists of 2 steps,
Calculating gradientandApplying to bayeisan parameters. - Before forward, network samples parameters with means & variances.
- Usually calling step function updates parameters, but not this case. After calling step function, you have to update bayesian parameters. Look at the ffg_model.py
- More benchmark cases
- Supports bayesian convolution
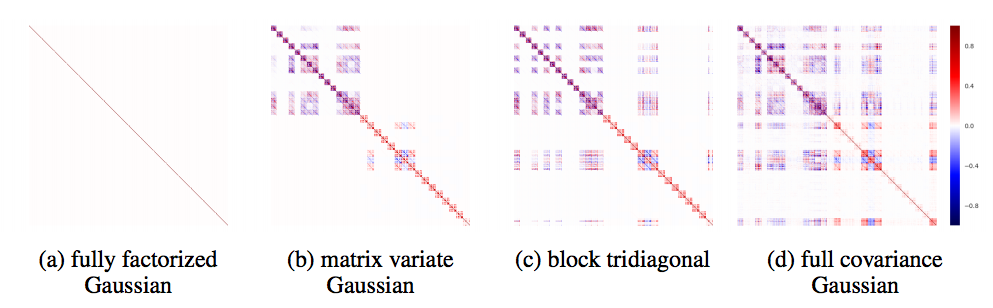
- Implement Block Tridiagonal Covariance, which is dependent between layers.
Visualization code(visualizer.py, utils.py) references to pytorch-CycleGAN-and-pix2pix(https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix) by Jun-Yan Zhu