NEWS: Version 0.6.9 (in development) adds a new cross-tabulation function, ctable(). To try it out, use devtools::install_github("dcomtois/summarytools", ref="dev"). It's a work-in-progress, but feedback is welcome.
Version 0.6.5 is now on CRAN, fixing several issues present in version 0.6. See README file for more details. The new version includes a vignette which complements the introduction on this page. You can find the vignette here: http://rpubs.com/dcomtois/summarytools_vignette
summarytools is an R package providing tools to neatly and quickly summarize data. Its main purpose is to provide hassle-free functions that every R programmer once wished were included in base R:
- frequency tables with proportions, cumulative proportions and missing data information.
- descriptive statistics with all common univariate statistics for numerical vectors
- dataframe summaries that facilitate data cleaning and firsthand evaluation
It also aims at making R a little easier to use for newcomers. With just a few lines of code, one can get a pretty good picture of the data at hand.
Newer versions (0.5 and above) support weights for freq() and descr(). Use devtools::install_github() to get the latest version (see How to install for detailed instructions).
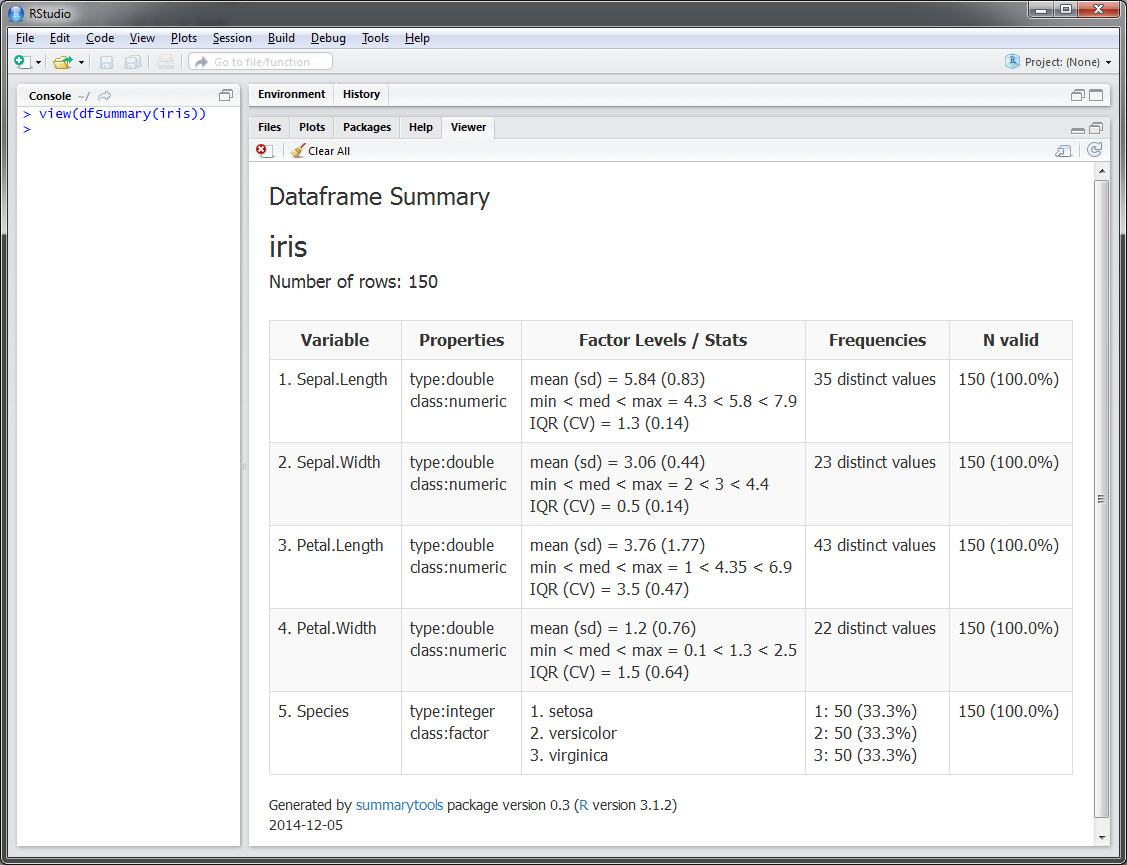
With just 2 lines of code, get a summary report of a dataframe, displayed directly in RStudio's Viewer pane:
> library(summarytools)
> view(dfSummary(iris))Building on the strengths of pander and htmltools, the summary reports produced by summarytools can be:
- Displayed as plain text in the R console
- Written to plain text files / markdown text files
- Written to html files that fire up in RStudio's Viewer pane or in your system's default browser.
- Support Hmisc and rapportools variable labels.
- Return table or dataframe objects for further manipulation if needed.
To install the latest stable version of summarytools, just type into your R console:
> install.packages("summarytools")For the most up-to-date version that has all the latest features but might also contain bugs, I invite you to first install the devtools package and then install summarytools through install_github():
> install.packages("devtools")
> library(devtools)
> install_github('dcomtois/summarytools')You can also see the source code and documentation on the official R site here.
The freq() function generates a table of frequencies with counts and percentages (including cumulative).
> library(summarytools)
> data(iris)
> # We'll insert some NA's for illustration purposes
> is.na(iris) <- matrix(sample(x = c(TRUE,FALSE), size = 150*5,
+ replace = T, prob = c(.1,.9)),nrow = 150)
> # and add a variable label for the Species column
> rapportools::label(iris$Species) <- "The Species (duh)"
> freq(iris$Species)
Dataframe: iris
Variable: Species
Label: The Species (duh)
Frequencies
N %Valid %Cum.Valid %Total %Cum.Total
---------------- --- -------- ------------ -------- ------------
setosa 46 33.58 33.58 30.67 30.67
versicolor 45 32.85 66.42 30 60.67
virginica 46 33.58 100 30.67 91.33
<NA> 13 NA NA 8.67 100
Total 150 100 100 100 100The descr() function generates common central tendency statistics and measures of dispersion for numerical data. It can handle single vectors as well as dataframes, in which case it just ignores non-numerical columns.
> data(iris)
> descr(iris)
Non-numerical variable(s) ignored: Species
Descriptive Univariate Statistics
Dataframe name: iris
Sepal.Length Sepal.Width Petal.Length Petal.Width
----------------- -------------- ------------- -------------- -------------
Mean 5.88 3.05 3.75 1.2
Std.Dev 0.84 0.44 1.76 0.77
Min 4.3 2 1 0.1
Max 7.9 4.4 6.9 2.5
Median 5.8 3 4.4 1.3
MAD 1.04 0.37 1.78 1.04
IQR 1.38 0.5 3.5 1.5
CV 6.98 6.99 2.13 1.56
Skewness 0.26 0.31 -0.3 -0.1
SE.Skewness 0.21 0.21 0.21 0.21
Kurtosis -0.68 0.17 -1.45 -1.37
Observations
Sepal.Length Sepal.Width Petal.Length Petal.Width
----------- -------------- ------------- -------------- -------------
Valid 134 (89.33%) 138 (92%) 134 (89.33%) 134 (89.33%)
<NA> 16 (10.67%) 12 (8%) 16 (10.67%) 16 (10.67%)
Total 150 150 150 150
If your eyes/brain prefer seeing things the other way around, just use "transpose=TRUE":
> descr(iris, transpose=TRUE)
Non-numerical variable(s) ignored: Species
Descriptive Statistics
Dataframe name: iris
Mean Std.Dev Min Max Median MAD IQR CV Skewness SE.Skewness Kurtosis
------------------ ------ --------- ----- ----- -------- ----- ----- ---- ---------- ------------- ----------
Sepal.Length 5.88 0.84 4.3 7.9 5.8 1.04 1.38 6.98 0.26 0.21 -0.68
Sepal.Width 3.05 0.44 2 4.4 3 0.37 0.5 6.99 0.31 0.21 0.17
Petal.Length 3.75 1.76 1 6.9 4.4 1.78 3.5 2.13 -0.3 0.21 -1.45
Petal.Width 1.2 0.77 0.1 2.5 1.3 1.04 1.5 1.56 -0.1 0.21 -1.37
Observations
Valid <NA> Total
------------------ ------------ ----------- -------
Sepal.Length 134 (89.33%) 16 (10.67%) 150
Sepal.Width 138 (92%) 12 (8%) 150
Petal.Length 134 (89.33%) 16 (10.67%) 150
Petal.Width 134 (89.33%) 16 (10.67%) 150
The dfSummary() function generates a table containing variable information (class(es) and type), common statistics for numerical data and frequency counts (as long as there are not too many distinct values -- and yes, you can specify the limit in the function call). Number and proportion of valid (non-missing) values are also reported, and variable labels can optionnaly be included.
> dfSummary(iris)----------------------------------------------------------------------------------------------
variable.name properties factor.levels.or.stats frequencies n.valid
--------------- ------------- --------------------------------- ------------------ -----------
Sepal.Length type:double mean (sd) = 5.88 (0.84) 35 distinct values 134 (89.3%)
class:numeric min < med < max = 4.3 < 5.8 < 7.9
IQR (CV) = 1.38 (0.14)
Sepal.Width type:double mean (sd) = 3.05 (0.44) 23 distinct values 138 (92.0%)
class:numeric min < med < max = 2 < 3 < 4.4
IQR (CV) = 0.5 (0.14)
Petal.Length type:double mean (sd) = 3.75 (1.76) 43 distinct values 134 (89.3%)
class:numeric min < med < max = 1 < 4.4 < 6.9
IQR (CV) = 3.5 (0.47)
Petal.Width type:double mean (sd) = 1.2 (0.77) 22 distinct values 134 (89.3%)
class:numeric min < med < max = 0.1 < 1.3 < 2.5
IQR (CV) = 1.5 (0.64)
Species type:integer 1. setosa 1: 46 (33.6%) 137 (91.3%)
class:factor 2. versicolor 2: 45 (32.8%)
3. virginica 3: 46 (33.6%)
----------------------------------------------------------------------------------------------Thanks to Gergely Daróczi's pander package, all functions can printout markdown; just use the option style="rmarkdown". That is useful for instance here on GitHub, where .md files are converted and displayed as html. Thanks to John MacFarlane's Pandoc, you can further convert markdown text files into a wide range of common formats such as .pdf, .docx and .odt, among others.
Here is an example of a markdown table, as processed by GitHub, using freq():
> freq(iris$Species, style="rmarkdown", plain.ascii=FALSE, missing="---")Dataframe name: iris
Variable name: Species
Variable label: The Species (duh)
Date: 2014-12-05
Frequencies
| N | %Valid | %Cum.Valid | %Total | %Cum.Total | |
|---|---|---|---|---|---|
| setosa | 46 | 33.58 | 33.58 | 30.67 | 30.67 |
| versicolor | 45 | 32.85 | 66.42 | 30 | 60.67 |
| virginica | 46 | 33.58 | 100 | 30.67 | 91.33 |
| <NA> | 13 | --- | --- | 8.67 | 100 |
| Total | 150 | 100 | 100 | 100 | 100 |
- We specified
plain.ascii=FALSE. This allows additional markup in the text (here, the bold-typed row names, added automatically by pander). - We used the option
missing="---", to show that if we don't like seeingNA's in our tables, it's quite easy to get rid of them or replace them with any character (or combination of characters).
To learn more about markdown and rmarkdown formats, see John MacFarlane's page and this RStudio's R Markdown Quicktour.
Version 0.5 of summarytools combines the strengths of the following packages and tools to generate basic html reports:
- RStudio's htmltools package
- The xtable package
- Bootstrap cascading stylesheets
When you become familiar with the method, You can achieve this in just one operation, but let's have a detailed walkthrough on how to generate and visualize an html report with summarytools.
- First, generate a summarytools object using one of
descr(),freq()ordfSummary():
> my.freq.table <- freq(iris$Species)- Next, use
print(), specifying themethodargument which can take one of the following values:method='browser'This creates an html report on-the-fly and makes it fire up in your system's default browser. The path to the report is returned.method='viewer'Same as "browser", except the report opens up in RStudio's Viewer pane (as demonstrated at the top of this page.)method='pander'This is the default value formethodand will not produce an html file. It will rather direct output to the console.
> print(my.freq.table, method="browser")- Since many of us like to stay in RStudio as much as possible, a wrapper function called
view()callsprint()specifyingmethod="viewer". You can stick toprint()altogether if you prefer.
There is another way to generate output right at the first function call to descr(), dfSummary() or freq(); it is to supply the argument "file" to any of those. For instance, the two following function calls will generate a markdown report, and then an html report from dfSummary():
> dfSummary(iris, style="grid", file="~/iris_dfSummary.md", escape.pipes=TRUE)
Output successfully written to file D:\Documents\iris_dfSummary.md
> dfSummary(iris, file="~/iris_dfSummary.html") # With html files, most of the other arguments are omitted.
Output successfully written to file D:\Documents\iris_dfSummary.htmlNote The "escape.pipes=TRUE" argument makes it so that Pandoc, in converting to alternative formats, handles correctly multiline cells in dfSummary() reports.
Some attributes attached to summarytools objects can be modified in order to change one of the elements displayed -- this is most usefull when generating html reports. In particular, you may want to change "df.name", "var.name" or "date". To do so, you would use R's attr() function in the following manner:
> attr(my.freq.table, "df.name") <- "The IRIS Dataframe"
> my.freq.table
Frequencies
Dataframe: The IRIS Dataframe
Variable: Species
N %Valid %Cum.Valid %Total %Cum.Total
---------------- --- -------- ------------ -------- ------------
setosa 50 33.33 33.33 33.33 33.33
versicolor 50 33.33 66.67 33.33 66.67
virginica 50 33.33 100 33.33 100
<NA> 0 NA NA 0 100
Total 150 100 100 100 100
When displaying summarytools objects in the console (as opposed to generating html reports), many other arguments can be specified so you get the format that you want. The most common are:
styleone of "simple" (default), "grid", "rmarkdown" and "multiline"justifyone of "left", "center", and "right"round.digitshow many decimals to show. This argument is also used for html reportsplain.asciiwhenTRUE, no markdown tags are used...and any of the other pander options
- Putting together the object's class(es), type (typeof), mode, storage mode, length, dim and object.size, all in a single table;
- Extending the
is()function in a way that the object is tested against all functions starting withis.-- see this post on StackOverflow for details; - Giving a list of the object's attributes names and length (c.g. rownames, dimnames, labels, etc.)
> what.is(c)
$properties
property value
1 class function
2 typeof builtin
3 mode function
4 storage.mode function
5 dim
6 length 1
7 object.size 56 Bytes
$extensive.is
[1] "is.function" "is.primitive" "is.recursive"
$function.type
[1] "primitive" "generic"
> what.is(NaN)
$properties
property value
1 class numeric
2 typeof double
3 mode numeric
4 storage.mode double
5 dim
6 length 1
7 object.size 48 Bytes
$extensive.is
[1] "is.atomic" "is.double" "is.na" "is.nan" "is.numeric" "is.vector"
$object.type
[1] "base"
Visit my professionnal site to learn more about what I do and services I offer: www.statconseil.com
The package comes with no guarantees. It is a work in progress and feedback / feature requests are most welcome. Just write me an email at dominic.comtois (at) gmail.com, or open an Issue if you find a bug.