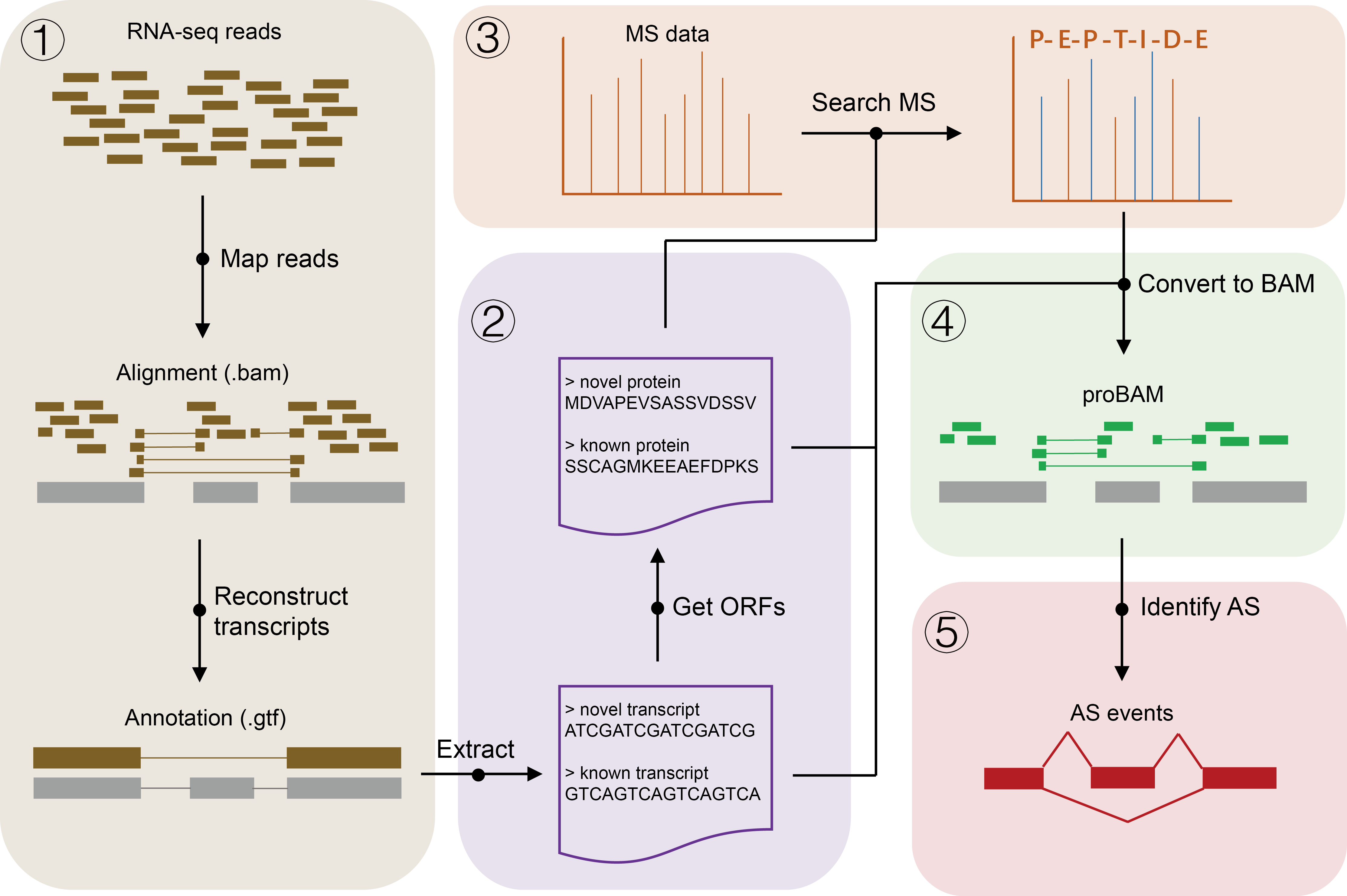
- Description: a Proteomics Alternative Splicing Screening pipeline.
-
Version: 1.1.0
-
Install
- unzip PASS-master.zip
- chmod a+x *
- export PATH=/PASS_install_path/:$PATH
-
System requirements
-
processRNASEQ:
-
Description: Align RNA-Seq reads to the reference genome and reconstruct transcripts.
-
Usage:
processRNASEQ [options] -g <genome> -f <.gtf> -r <.fastq>-g Genome bowtie2 index name. -f Gene annotation file, .gtf format. -r File names for sequencing reads, .fastq format. - Compressed files (.fastq.gz) are also supported. - Paired-end files separated by commas. -t Path to tophat, eg. /home/user/bin/tophat - By default, we try to search tophat in system PATH. -c Path to cufflinks, eg. /home/user/bin/cufflinks - By default, we try to search cufflinks in system PATH. -p Number of used threads. [Default: 12] -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
processRNASEQ -g path_genomeandbowtie2index/genome.test -f exampleData/genes.test.gtf -r exampleData/Sample_R1.fastq.gz,exampleData/Sample_R2.fastq.gz
-
-
getORF:
-
Description: Protein sequences translation.
-
Usage:
getORF [options] -f <.gtf> -g <genome.fa>-f File name of gene annotation, .gtf format. - Recommend cufflinks to generate this file. -g Reference genome file name, fasta format. -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
getORF -f exampleData/transcripts.gtf -g exampleData/genome.test.fa -
Output:
- transcripts.longestorf.gtf
- transcript.longestorf.fa
- protein.longestorf.fa
-
-
searchMS:
-
Description: Search MS file against protein sequence database.
-
Usage:
searchMS [options] -s <MSGF_path> -m <example.mzML> -f <protein.fa>-s Path to MSGFPlus.jar. eg. ~/software/MSGF. -m MS/MS file. - Support file formats including .mzML, .mzXML, .mgf, .ms2, .pkl and _dta.txt - Spectral should be centroided. -f Protein sequences -p Number of used threads. [Default: 12] -t Modification file name. -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
searchMS -s ~/software/MSGF -m exampleData/example.mzML -f exampleData/protein.longestorf.fa -
Output:
- PSM.tab
-
-
generateSAM:
-
Description: Convert peptide spectal matches to alignment file.
-
Usage:
generateSAM [options] -m <PSM> -f <.gtf> -t <transcript.fa> -p <protein.fa>-m Peptide spectral matches. -f File name of gene annotation, .gtf format. -t File name of transcript sequences, .fa format. -p File name of protein sequences, .fa format. -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
generateSAM -m exampleData/PSM.tab -f exampleData/transcripts.longestorf.gtf -t exampleData/transcript.longestorf.fa -p exampleData/protein.longestorf.fa -
Output:
- PSM.sam
-
-
screenAS:
-
Description: Detect AS events from annotation and alignment file.
-
Note: This function code is sourced from MATS.
-
Usage:
screenAS [options] -s <PSM.sam> -g <genes.gtf>-s Sam format file generated by proteome identification. -g Gene annotation file, .gtf format. -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
screenAS -s exampleData/PSM.sam -g exampleData/transcripts.longestorf.gtf -
Output
- summary.txt
- PASS.SE.txt
- PASS.RI.txt
- PASS.MXE.txt
- PASS.A5SS.txt
- PASS.A3SS.txt
- PASS.AFE.txt
- PASS.ALE.txt
-
-
PASS:
-
Description: All-in-one command.
-
Usage:
PASS [options] -g <genome> -f <genes.gtf> -r <reads.fastq> -s <MSGFPlus.jar> -m <example.mzML>-g Genome bowtie2 index name. -f Gene annotation file, .gtf format. -r File names for sequencing reads, .fastq format. - Compressed files (.fastq.gz) are also supported. - Paired-end files separated by commas. -t Path to tophat, eg. /home/user/bin/tophat - By default, we try to search tophat in system PATH. -c Path to cufflinks, eg. /home/user/bin/cufflinks - By default, we try to search cufflinks in system PATH. -p Number of used threads. [Default: 12] -s Path to MSGFPlus.jar. eg. ~/software/MSGF. -m MS/MS file. - Support file formats including .mzML, .mzXML, .mgf, .ms2, .pkl and _dta.txt - Spectra should be centroided. -d Modification file name. -o Output folder. [Default: ./PASS_out] -h Help message. -
Example:
PASS -g path_genomeandbowtie2index/genome.test -f exampleData/genes.test.gtf -r exampleData/Sample_R1.fastq.gz,exampleData/Sample_R2.fastq.gz -s ~/software/MSGF -m exampleData/example.mzML -p 4 -
Output:
- summary.txt
- PASS.SE.txt
- PASS.RI.txt
- PASS.MXE.txt
- PASS.A5SS.txt
- PASS.A3SS.txt
- PASS.AFE.txt
- PASS.ALE.txt
-
-
Contact:
Peng Wu; wupeng1@ihcams.ac.cn