This repository holds Docker build scripts, workflows and benchmarks for variant calling used by the Human Pangenome Project.
Reads/assemblies are aligned to the following reference genomes:
-
GRCh38_no_alt: GRCh38 without ALT contigs$ wget https://s3-us-west-2.amazonaws.com/human-pangenomics/working/HPRC_PLUS/GRCh38/assemblies/GCA_000001405.15_GRCh38_no_alt_analysis_set.fna.gz $ gunzip GCA_000001405.15_GRCh38_no_alt_analysis_set.fna.gz # pbsv doesn't recognize *.fna $ mv GCA_000001405.15_GRCh38_no_alt_analysis_set.fna GCA_000001405.15_GRCh38_no_alt_analysis_set.fa -
CHM13Y_EBV: CHM13 v1.1 + chrY and chrEBV fromGRCh38_no_alt$ wget https://g-f20d48.ad0e3.03c0.data.globus.org/CHM13Y_EBV_v1.1.fasta.gz $ gunzip CHM13Y_EBV_v1.1.fasta.gz
-
Compute high frequency k-mers in a reference genome using meryl
#GRCh38_no_alt $ meryl count k=15 output merylDB GCA_000001405.15_GRCh38_no_alt_analysis_set.fa $ meryl print greater-than distinct=0.9998 merylDB > GCA_000001405.15_GRCh38_no_alt_analysis_set_repetitive_k15.txt #CHM13Y_EBV $ meryl count k=15 output merylDB CHM13Y_EBV_v1.1.fasta $ meryl print greater-than distinct=0.9998 merylDB > CHM13Y_EBV_v1.1_repetitive_k15.txt
-
For each movie, align the HiFi reads using Winnowmap v2.03 and sort alignments by coordinates using SAMtools
$ winnowmap -W ${MERYLFILE} -t ${THREADS} \ -R "@RG\tID:${RGID}\tPL:PACBIO\tDS:READTYPE=CCS\tPU:${MOVIE}\tSM:${SAMPLE}\tPM:SEQUEL" \ -x map-pb -a -Y -L --eqx --cs ${REFFASTA} ${FASTQ} \ | samtools sort -m4G -@ ${THREADS} -o ${SAMPLE}.${MOVIE}.${REF}.bam
-
Merge alignments, add MD tags, and index aligments using SAMtools
$ samtools merge -@ ${THREADS} ${SAMPLE}.${REF}.merged.bam ${BAMFILES} \ && samtools calmd -b -@ ${THREADS} ${SAMPLE}.${REF}.merged.bam ${REFFASTA} > ${SAMPLE}.${REF}.bam \ && samtools index -@ ${THREADS} ${SAMPLE}.${REF}.bam
-
Compute high frequency k-mers in a reference genome using meryl
#GRCh38_no_alt $ meryl count k=15 output merylDB GCA_000001405.15_GRCh38_no_alt_analysis_set.fa $ meryl print greater-than distinct=0.9998 merylDB > GCA_000001405.15_GRCh38_no_alt_analysis_set_repetitive_k15.txt #CHM13Y_EBV $ meryl count k=15 output merylDB CHM13Y_EBV_v1.1.fasta $ meryl print greater-than distinct=0.9998 merylDB > CHM13Y_EBV_v1.1_repetitive_k15.txt
-
For each flow cell, align the ONT reads using Winnowmap v2.03 and sort alignments by coordinates using SAMtools
$ winnowmap -W ${MERYLFILE} -t ${THREADS} \ -R "@RG\tID:${RGID}\tPL:ONT\tSM:${SAMPLE}" \ -x map-ont -a -Y -L --eqx --cs ${REFFASTA} ${FASTQ} \ | samtools sort -m4G -@ ${THREADS} -o ${RGID}.${REF}.bam
-
Merge alignments, add MD tags, and index aligments using SAMtools
$ samtools merge -@ ${THREADS} ${SAMPLE}.${REF}.merged.bam ${BAMFILES} \ && samtools calmd -b -@ ${THREADS} ${SAMPLE}.${REF}.merged.bam ${REFFASTA} > ${SAMPLE}.${REF}.bam \ && samtools index -@ ${THREADS} ${SAMPLE}.${REF}.bam
Align paternal and maternal assemblies using minimap2 v2.21
# paternal
$ minimap2 -x asm5 -a --eqx --cs -t ${THREADS} ${REFFASTA} ${SAMPLE}.paternal.f1_assembly_v2_genbank.fa \
| samtools sort --write-index -m4G -@ ${THREADS} -o ${SAMPLE}.paternal.${REF}.bam##idx##${SAMPLE}.paternal.${REF}.bam.bai
# maternal
$ minimap2 -x asm5 -a --eqx --cs -t ${THREADS} ${REFFASTA} ${SAMPLE}.maternal.f1_assembly_v2_genbank.fa \
| samtools sort --write-index -m4G -@ ${THREADS} -o ${SAMPLE}.maternal.${REF}.bam##idx##${SAMPLE}.maternal.${REF}.bam.baiDocker image: google/deepvariant:1.1.0
$ run_deepvariant --model_type PACBIO \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
--reads hifi_alignments/HG002.GRCh38_no_alt.bam \
--output_gvcf calls/deepvariant/HG002.GRCh38_no_alt.deepvariant1.g.vcf.gz \
--output_vcf calls/deepvariant/HG002.GRCh38_no_alt.deepvariant1.vcf.gz \
--num_shards 16 \
--call_variants_extra_args "use_openvino=true"Docker image: wwliao/hpp_whatshap:1.0.0--4423f2ed9c728c3e569482b5693006e467a3a596
$ whatshap phase --output calls/deepvariant/HG002.deepvariant1.phased.vcf.gz \
--reference GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
calls/deepvariant/HG002.GRCh38_no_alt.deepvariant1.vcf.gz \
hifi_alignments/HG002.GRCh38_no_alt.bam \
&& bcftools index -t calls/deepvariant/HG002.deepvariant1.phased.vcf.gz \
&& whatshap haplotag --output calls/deepvariant/HG002.GRCh38_no_alt.haplotagged.bam \
--reference GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
calls/deepvariant/HG002.deepvariant1.phased.vcf.gz \
hifi_alignments/HG002.GRCh38_no_alt.bam \
&& samtools index calls/deepvariant/HG002.GRCh38_no_alt.haplotagged.bamDocker image: google/deepvariant:1.1.0
$ run_deepvariant --model_type PACBIO \
--ref GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
--reads calls/deepvariant/HG002.GRCh38_no_alt.haplotagged.bam \
--use_hp_information \
--output_gvcf calls/deepvariant/HG002.GRCh38_no_alt.deepvariant.g.vcf.gz \
--output_vcf calls/deepvariant/HG002.GRCh38_no_alt.deepvariant.vcf.gz \
--num_shards 16 \
--call_variants_extra_args "use_openvino=true"To increase pbsv calling performance, it is recommended to provide a tandem repeat annotation in BED format.
Docker image: wwliao/hpp_pbsv:1.0.0--5ac93f8f6bd021a33feeb3fbca50466f27847b15
$ pbsv discover -s HG002 \
--tandem-repeats GRCh38_no_alt.trf.bed \
hifi_alignments/HG002.GRCh38_no_alt.bam \
calls/pbsv/outputs/HG002.GRCh38_no_alt.svsig.gz \
&& pbsv call --ccs \
--preserve-non-acgt \
-t DEL,INS,INV,DUP,BND \
-m 40 \
-j 12 \
GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
calls/pbsv/outputs/HG002.GRCh38_no_alt.svsig.gz \
calls/pbsv/HG002.GRCh38_no_alt.pbsv.vcf \
&& bcftools sort -m4G \
-Oz \
-o calls/pbsv/HG002.GRCh38_no_alt.pbsv.vcf.gz \
calls/pbsv/HG002.GRCh38_no_alt.pbsv.vcf \
&& bcftools index -t calls/pbsv/HG002.GRCh38_no_alt.pbsv.vcf.gz \
&& rm calls/pbsv/HG002.GRCh38_no_alt.pbsv.vcfDocker image: wwliao/hpp_sniffles:1.0.0--de39905b020c976bbebdd99d32c5df9b2c812ab4
$ sniffles -s 4 \
-l 40 \
-n -1 \
--cluster \
--ccs_reads \
-t 12 \
-m hifi_alignments/HG002.GRCh38_no_alt.bam \
-v calls/sniffles/outputs/HG002.GRCh38_no_alt.sniffles.raw.vcf \
&& echo HG002 > HG002.txt \
&& bcftools reheader -s HG002.txt \
-o calls/sniffles/outputs/HG002.GRCh38_no_alt.sniffles.unrefined.vcf \
calls/sniffles/outputs/HG002.GRCh38_no_alt.sniffles.raw.vcf \
&& rm HG002.txt calls/sniffles/outputs/HG002.GRCh38_no_alt.sniffles.raw.vcf \
&& iris genome_in=GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
vcf_in=calls/sniffles/outputs/HG002.GRCh38_no_alt.sniffles.unrefined.vcf \
reads_in=hifi_alignments/HG002.GRCh38_no_alt.bam \
vcf_out=calls/sniffles/HG002.GRCh38_no_alt.sniffles.vcf \
threads=12 \
out_dir=calls/sniffles/tmp/HG002 \
--also_deletions \
--hifi \
--rerunracon \
--keep_long_variants \
&& bcftools sort -m4G \
-Oz \
-o calls/sniffles/HG002.GRCh38_no_alt.sniffles.vcf.gz \
calls/sniffles/HG002.GRCh38_no_alt.sniffles.vcf \
&& bcftools index -t calls/sniffles/HG002.GRCh38_no_alt.sniffles.vcf.gz \
&& rm -rf calls/sniffles/tmp/HG002 \
&& rm calls/sniffles/HG002.GRCh38_no_alt.sniffles.vcfDocker image: wwliao/hpp_svim:1.0.0--93e7668e2ecd1fc315126e69da862fd365339509
$ svim alignment --interspersed_duplications_as_insertions \
--read_names \
--zmws \
--cluster_max_distance 0.5 \
--minimum_depth 4 \
--min_sv_size 40 \
--sample HG002 \
calls/svim/outputs/HG002 \
hifi_alignments/HG002.GRCh38_no_alt.bam \
GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
&& bcftools sort -Oz \
-o calls/svim/HG002.GRCh38_no_alt.svim.vcf.gz \
calls/svim/outputs/HG002/variants.vcf \
&& bcftools index -t calls/svim/HG002.GRCh38_no_alt.svim.vcf.gzDocker image: wwliao/hpp_svimasm:1.0.0--896384d810154ae24154f2143fe327e9e57b326a
$ svim-asm diploid --interspersed_duplications_as_insertions \
--query_names \
--min_sv_size 40 \
--sample HG002 \
calls/svim-asm/outputs/HG002 \
asm_alignments/HG002.paternal.GRCh38_no_alt.bam \
asm_alignments/HG002.maternal.GRCh38_no_alt.bam \
GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
&& bcftools sort -Oz \
-o calls/svim-asm/HG002.GRCh38_no_alt.svim-asm.vcf.gz \
calls/svim-asm/outputs/HG002/variants.vcf \
&& bcftools index -t calls/svim-asm/HG002.GRCh38_no_alt.svim-asm.vcf.gzWe compare our HG002 SV callsets with the GIAB v0.6 Tier 1 SV benchmark set for HG002. Since the GIAB benchmark set is on GRCh37, we lifted over it to GRCh38:
| Caller | Recall % | Precision % | F1 % | GT-Recall % | GT-Precision % | GT-F1 % |
|---|---|---|---|---|---|---|
| pbsv | 95.57 | 87.87 | 91.56 | 95.32 | 82.88 | 88.66 |
| Sniffles + Iris | 95.29 | 93.03 | 94.15 | 90.28 | 42.75 | 58.02 |
| SVIM | 97.14 | 70.87 | 81.95 | 97.01 | 67.75 | 79.79 |
| SVIM (score >= 10) |
93.43 | 92.26 | 92.85 | 93.24 | 89.38 | 91.27 |
| SVIM-asm | 97.30 | 90.84 | 93.96 | 96.16 | 63.06 | 76.17 |
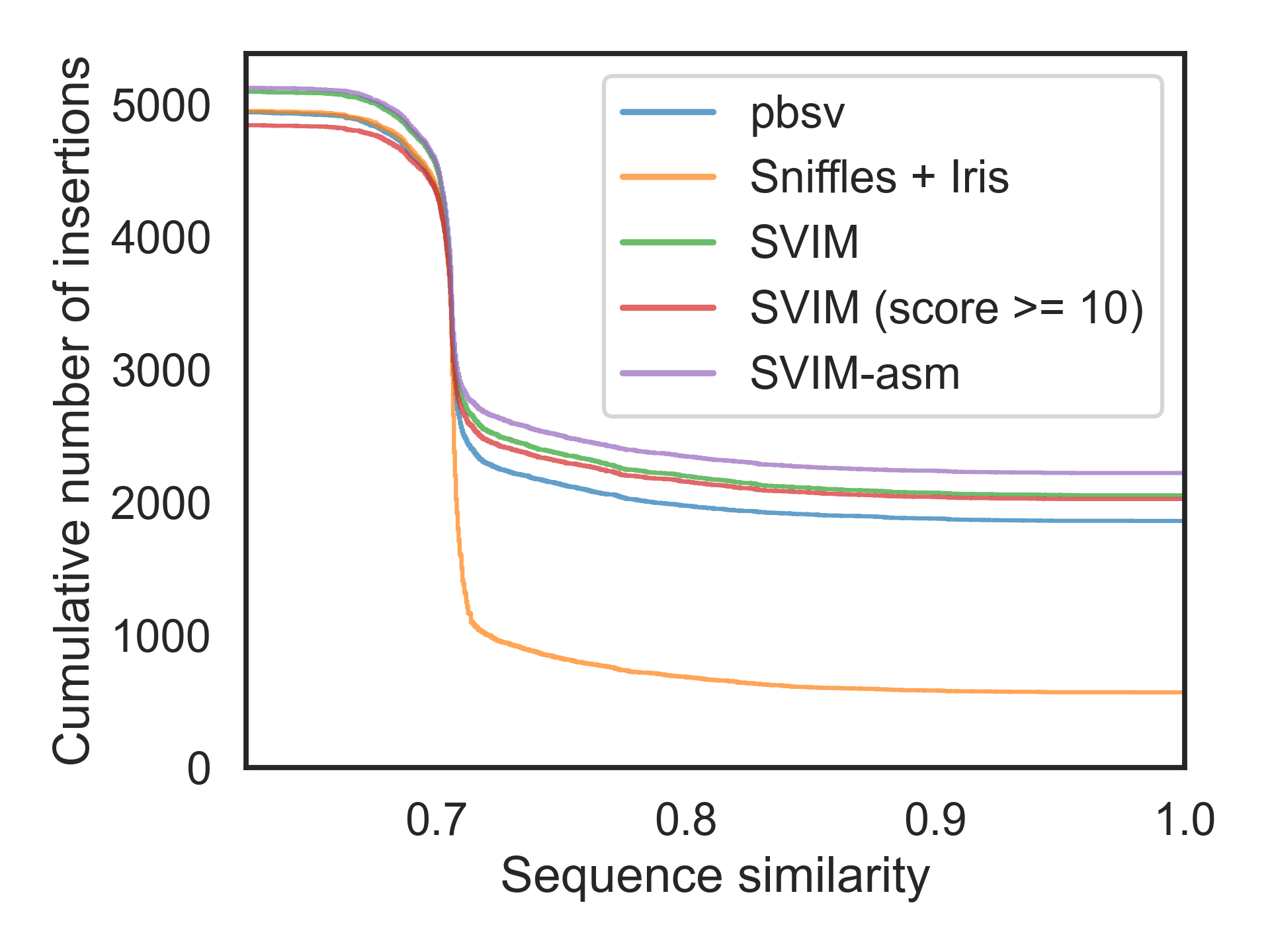
Figure 1. Insertion Sequence Similarity
Note that the sequence similarity is defined as Levenshtein distance ratio.
$ cat <(zcat HG002.GRCh38_no_alt.pbsv.vcf.gz | grep "^#") \
<(zcat HG002.GRCh38_no_alt.pbsv.vcf.gz | grep -vE "^#" | grep 'INS\|DEL') \
| bgzip -c > HG002.GRCh38_no_alt.pbsv.filtered.vcf.gz \
&& bcftools index -t HG002.GRCh38_no_alt.pbsv.filtered.vcf.gz$ cat <(zcat HG002.GRCh38_no_alt.sniffles.vcf.gz | grep "^#") \
<(zcat HG002.GRCh38_no_alt.sniffles.vcf.gz | grep -vE "^#" | grep 'INS\|DEL') \
| bgzip -c > HG002.GRCh38_no_alt.sniffles.filtered.vcf.gz \
&& bcftools index -t HG002.GRCh38_no_alt.sniffles.filtered.vcf.gz$ cat <(zcat HG002.GRCh38_no_alt.svim.vcf.gz | grep "^#") \
<(zcat HG002.GRCh38_no_alt.svim.vcf.gz | grep -vE "^#" | grep 'INS\|DEL' \
| awk '{ if($6>='${SCORE}') { print $0 } }') \
| bgzip -c > HG002.GRCh38_no_alt.svim.filtered.vcf.gz \
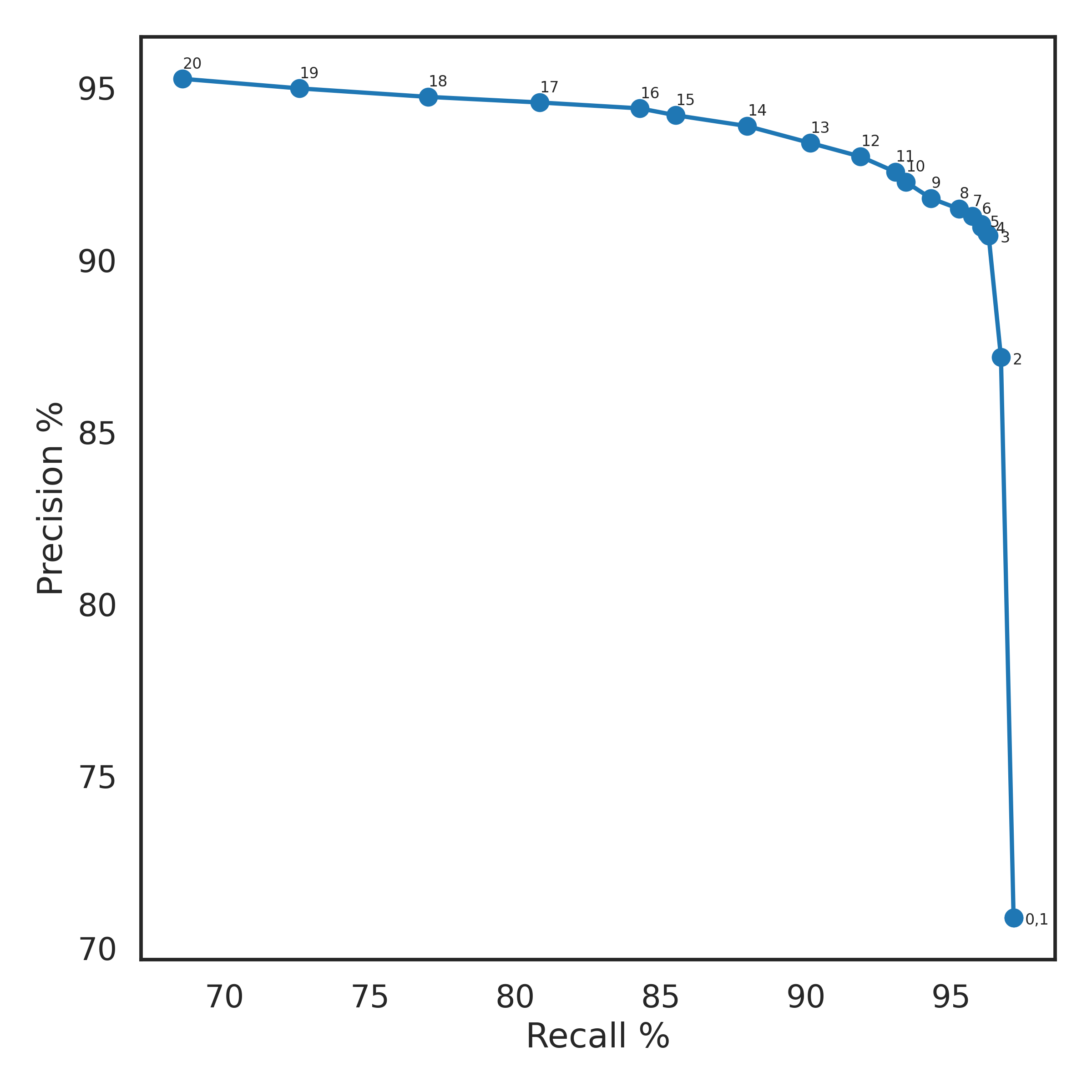
&& bcftools index -t HG002.GRCh38_no_alt.svim.filtered.vcf.gzFigure 2. SVIM Precision-Recall Curve
$ cat <(zcat HG002.GRCh38_no_alt.svim-asm.vcf.gz | grep "^#") \
<(zcat HG002.GRCh38_no_alt.svim-asm.vcf.gz | grep -vE "^#" | grep 'INS\|DEL') \
| bgzip -c > HG002.GRCh38_no_alt.svim-asm.filtered.vcf.gz \
&& bcftools index -t HG002.GRCh38_no_alt.svim-asm.filtered.vcf.gzDocker image: wwliao/hpp_truvari:1.0.0--9352c3b9ffbe4680b1712ee465acdaec7f6f909f
$ truvari bench -f GCA_000001405.15_GRCh38_no_alt_analysis_set.fa \
-b HG002_SVs_Tier1_v0.6.GRCh38.vcf.gz \
-c HG002.GRCh38_no_alt.${CALLER}.filtered.vcf.gz \
--includebed HG002_SVs_Tier1_v0.6.GRCh38.bed \
-o bench-${CALLER} \
-r 1000 \
-p 0.01 \
--passonly \
--giabreport