Introduction
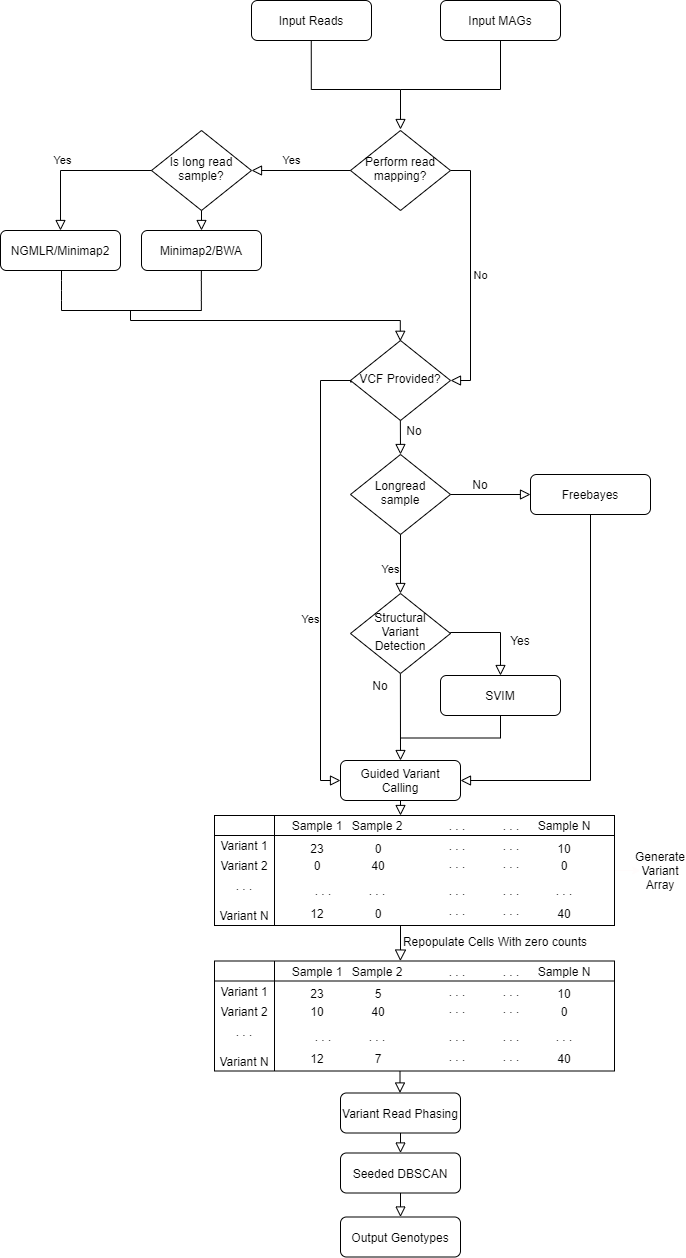
Lorikeet is a within-species variant analysis pipeline for metagenomic communities that utilizes both long and short read datasets. Lorikeet combines short read variant calling with Freebayes and long read structural variant calling with SVIM to generate a complete variant landscape present across samples within a microbial community. SNV are also linked together using read information to help provide likely genotypes based on observed physical linkages.
Installation
Option 1: Conda - Recommended
Install into current conda environment:
conda install lorikeet-genome
Create fresh conda environment and install lorikeet there:
conda create -n lorikeet -c bioconda lorikeet-genome && \
conda activate lorikeet
Option 2: Cargo
conda create -n lorikeet -y -c conda-forge -c bioconda -c defaults -y parallel pysam=0.16 svim \
freebayes=1.3.2 prokka samtools bcftools vt rust clangdev pkg-config zlib gsl starcode openblas bwa minimap2 \
fastani dashing r-base && \
conda activate lorikeet && \
cargo install lorikeet-genome
Option 3: Build manually
You may need to manually set the paths for C_INCLUDE_PATH, LIBRARY_PATH, LIBCLANG_PATH, and OPENSSL_DIR to their corresponding
paths in the your conda environment if they can't properly be found on your system.
conda create -n lorikeet -y -c conda-forge -c bioconda -c defaults -y parallel pysam=0.16 svim \
freebayes=1.3.2 prokka samtools bcftools vt rust clangdev pkg-config zlib gsl starcode openblas bwa minimap2 \
fastani dashing r-base && \
conda activate lorikeet && \
git clone https://github.com/rhysnewell/Lorikeet/git && \
cd Lorikeet && \
bash build.sh
Usage
Input can either be reads and reference genome, or MAG. Or a BAM file and associated genome.
Strain genotyping analysis for metagenomics
Usage: lorikeet <subcommand> ...
Main subcommands:
genotype *Experimental* Resolve strain-level genotypes of MAGs from microbial communities
polymorph Calculate variants along contig positions
summarize Summarizes contig stats from multiple samples
evolve Calculate dN/dS values for genes from read mappings
Less used utility subcommands:
kmer Calculate kmer frequencies within contigs
filter Remove (or only keep) alignments with insufficient identity
Other options:
-V, --version Print version information
Rhys J. P. Newell <r.newell near uq.edu.au>
Genotype from bam:
lorikeet genotype -b input.bam -r input_genome.fna --e-min 0.1 --e-max 0.5 --pts-min 0.1 --pts-max 0.5
Genotype from short reads and longread bam:
lorikeet genotype -r input_genome.fna -1 forward_reads.fastq -2 reverse_reads.fastq -l longread.bam
Workflow
Output
Genotype
Genotype will produce multiple .fna files representative of the expected strain level genotypes
Polymorph
Polymorph produces a tab delimited file containing possible variants and their positions within the reference
Evolve
Evolve will produce dN/dS values within coding regions based on the possible variants found along the reference. These dN/dS values only take single nucleotide polymorphisms into account but INDELs can still be reported.
Summarize
Produce a SNP summary report for each contig in the provided MAG