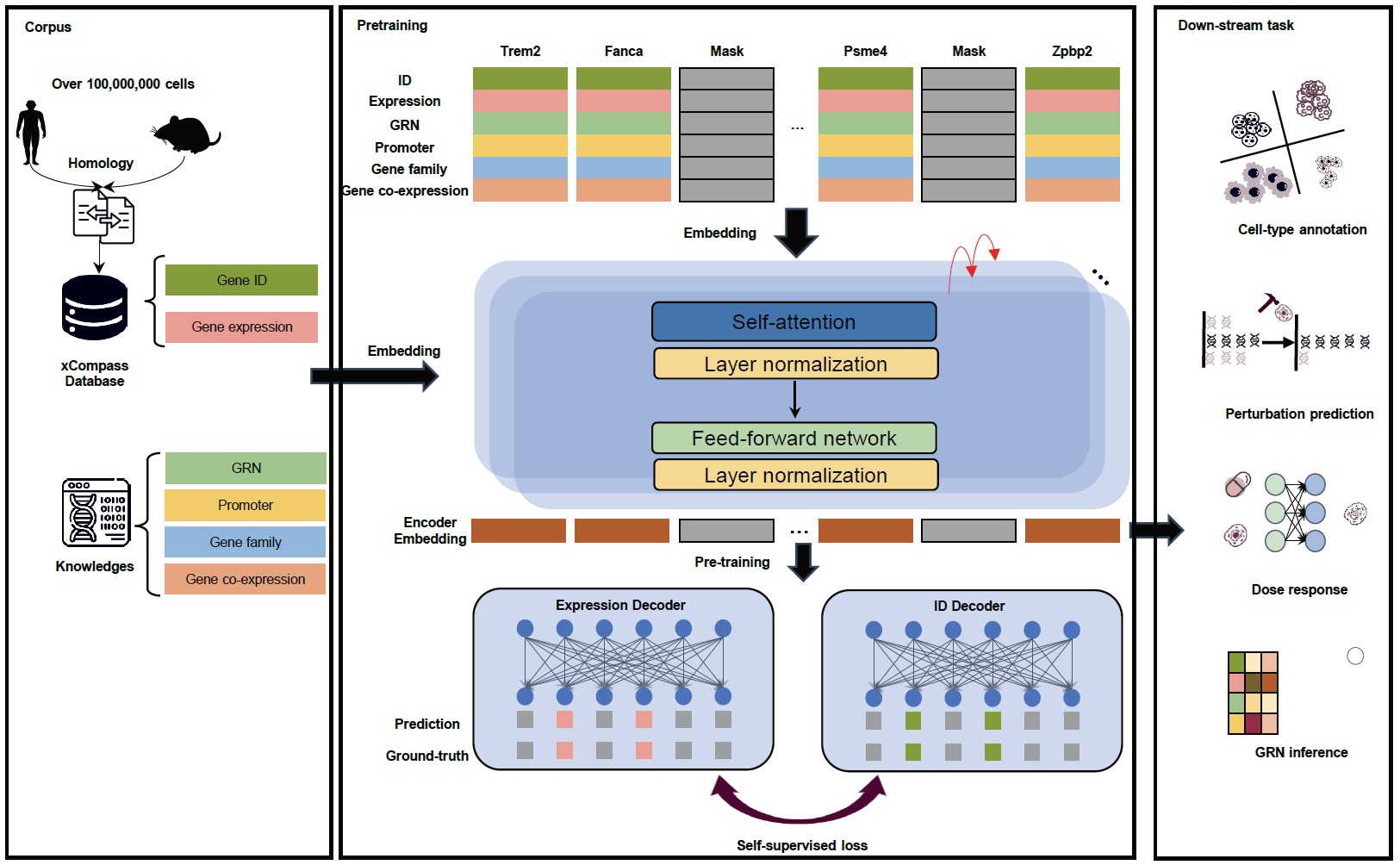
Deciphering universal gene regulatory mechanisms in diverse organisms holds great potential for advancing our knowledge of fundamental life processes and facilitating clinical applications. However, the traditional research paradigm primarily focuses on individual model organisms and does not integrate various cell types across species. Recent breakthroughs in single-cell sequencing and deep learning techniques present an unprecedented opportunity to address this challenge. In this study, we developed GeneCompass, a knowledge-informed cross-species foundation model, pre-trained on an extensive dataset of over 120 million human and mouse single-cell transcriptomes. During pre-training, GeneCompass effectively integrated four types of prior biological knowledge to enhance our understanding of gene regulatory mechanisms in a self-supervised manner. By fine-tuning for multiple downstream tasks, GeneCompass outperformed state-of-the-art models in diverse applications for a single species and unlocked new realms of cross-species biological investigations. We also employed GeneCompass to search for key factors associated with cell fate transition and showed that the predicted candidate genes could successfully induce the differentiation of human embryonic stem cells into the gonadal fate. Overall, GeneCompass demonstrates the advantages of using artificial intelligence technology to decipher universal gene regulatory mechanisms and shows tremendous potential for accelerating the discovery of critical cell fate regulators and candidate drug targets.
- This is the official repository of Genecompass which provides training&finetuning code and pretraining checkpoints.
- GeneCompass is implemented based on Pytorch. We use pytorch-1.13.1 and cuda-11.7. Other version could be also compatible. Building the environment and installing needed package.
- First, you should add GeneCompass main folder to the system path, and install requried packages. Run the following in shell:
nano ~/.bashrc
# Add to the file
export PATH="$PROJECT_DIR:$PATH"
pip install -r requirements.txt
- Optional you can use setup.sh to install GeneCompass automatically:
cd /path/to/genecompass
chmod +x setup.sh
./setup.sh
source ~/.bashrc
- [Optional] We recommend using wandb for logging and visualization.
pip install wandb
Pretrained models of GeneCompass on 100 million single-cell transcriptomes from humans and mice. Put pretrained_model dir under main path.('./pretrained_models/GeneCompass_Small', './pretrained_models/GeneCompass_Base')
| Model | Description | Download |
|---|---|---|
| GeneCompass_Small | Pretrained on 6-layer GeneCompass. | Link |
| GeneCompass_Base | Pretrained on 12-layer GeneCompass. | Link |
We here show the data processing procedures with preprocess.
GeneCompass utilizes over 100 million single-cell transcriptomes from humans and mice. We provide 50K, 500k and 5M pretrained data of human and mouse respectively. You can download and put dataset dir under main path.(e.g. './data/genecompass_5M/')
| Data | Description | Download |
|---|---|---|
| 0.05M | Pretrained data of 50K single cells. | Link-Human Link-Mouse |
| 0.5M | Pretrained data of 500k single cells. | Link-Human Link-Mouse |
| 5M | Pretrained data of 5M single cells. | Link-Human Link-Mouse |
For single-species cell-type annotation tasks, GeneCompass was conducted in different human datasets, i.e., multiple sclerosis (hMS), lung (hLung) and liver (hLiver), and diverse mouse datasets, i.e., brain (mBrain), lung (mLung) and pancreas (mPancreas).
We provide preprocessed data for above datasets here, and you only need to download the dataset and put dataset dir under main path.(e.g. './data/cell_type_annotation/hMS')
| Dataset | Description | Source | Download |
|---|---|---|---|
| hMS | Multiple sclerosis from human. | EMBL-EBI | Link |
| hLung | Lung from human. | GEO: GSE136831 | Link |
| hLiver | Liver from human. | Sharma et al | Link |
| mBrain | Brain from mouse. | GEO: GSE224407 | Link |
| mLung | Lung from mouse. | GEO: GSE225664 | Link |
| mPancreas | Pancreas from mouse. | GEO: GSE132188 | Link |
GEO means Gene Expression Omnibus.
Here we provided an example script, run by:
cd examples/
bash run_pretrain_genecompass_w_human_mouse_100M.sh
or you can just run the below as an example.
cd examples/
export CUDA_VISIBLE_DEVICES=0,1,2,3,4,5,6,7
python -m torch.distributed.launch --nproc_per_node= 8 \
--nnodes=1 \
--node_rank=0 \
--master_port=12348 \
pretrain_genecompass_w_human_mouse_base.py \
--run_name="test" \
--seed_num=0 \
--seed_val=42 \
--token_dict_path="../prior_knowledge/human_mouse_tokens.pickle" \
--dataset_directory="/home/share/genecompass_github/xCompass/data/6000W_control_lung_human" \
--num_train_epochs=5 \
--train_micro_batch_size_per_gpu=10 \
--max_learning_rate=5e-5 \
--warmup_steps=10000 \
--emb_warmup_steps=10000 \
--lr_scheduler_type="linear" \
--weight_decay=0.01 \
--dataloader_num_workers=0 \
--output_directory="./outputs" \
--do_train \
--save_model \
--save_strategy="steps" \
--save_steps=100000 \
--fp16 \
We performed a comprehensive analysis of diverse organ datasets from humans and mice. See cell-type annotation example on hMS.
The GRN prediction is based on the cosine similarity of gene embeddings between origin state and in silico perturbed state. By comparing the cosine similarity among genes except for the TF, those with low cosine similarity genes are prone to be considered as Target Genes (TG). See insilico_perturbation example.
Part here is using Gears to implement large model coding to predict changes in gene expression after gene perturbation in downstream tasks. The overall code is based on the initial Gears, where the parts that generate the code are tweaked and modified.(Gears: Implements the predictive function of Gears expression change.) Data preprocessing code: A layer supplement to the data set that is required for Gears-specific modifications, outside of the genecompass large model. Can be directly by example corresponding gears in the environment for the server
This task is included in GRN inference folder, please go to corresponding folder and read README.md to implement the task
This task is included in Drug dose response folder, please go to corresponding folder and read README.md to implement the task
This task is included in Gene expression profiling folder, please go to corresponding folder and read README.md to implement the task
If you find this code useful for your research, please consider citing:
@article{yang2023genecompass,
title={Genecompass: Deciphering universal gene regulatory mechanisms with knowledge-informed cross-species foundation model},
author={Yang, Xiaodong and Liu, Guole and Feng, Guihai and Bu, Dechao and Wang, Pengfei and Jiang, Jie and Chen, Shubai and Yang, Qinmeng and Zhang, Yiyang and Man, Zhenpeng and others},
journal={bioRxiv},
pages={2023--09},
year={2023},
publisher={Cold Spring Harbor Laboratory}
}