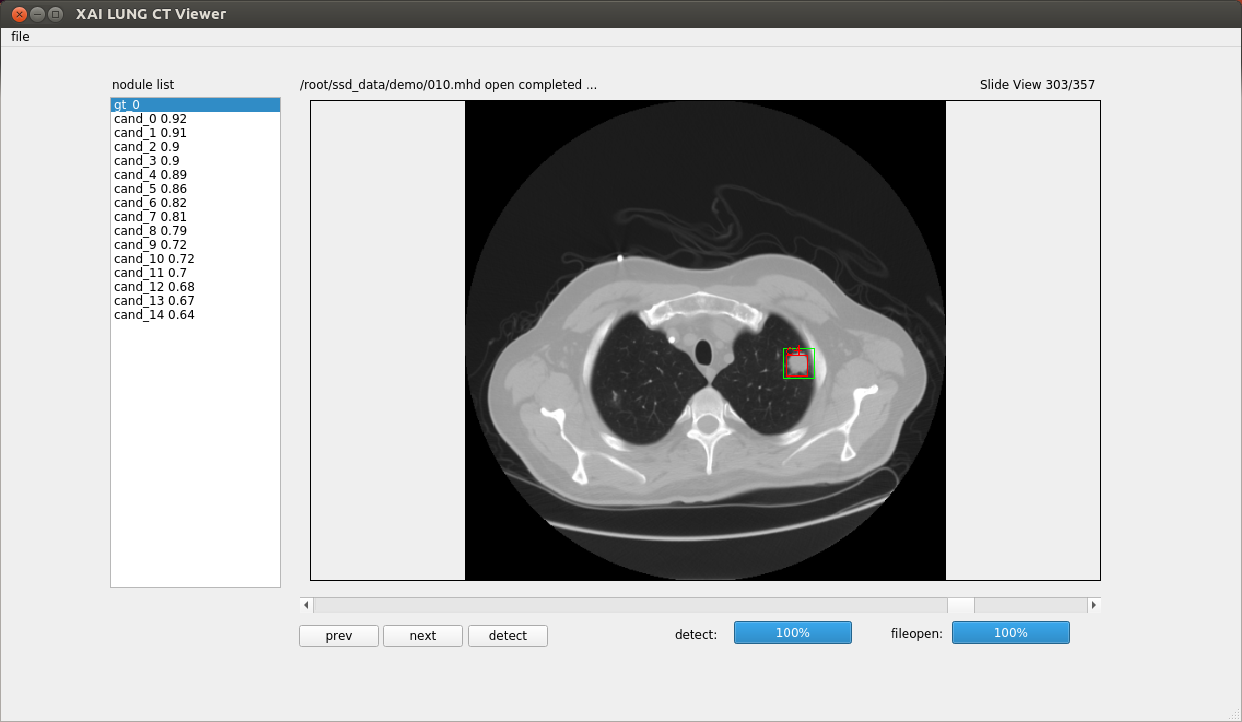
lung nodule detector using 3d resnet
python 3.5.2
pytorch 0.2.0
pyqt 5
docker pull likebullet86/luna16_detector
- settings (detector_viewer/xai_viewer.py)
- self.init_openpath
- self.label_dirpath
- self.detect_resume
- self.gpu
- excute
- python xai_viewer.py
- for using viewer in docker
- xhost + (excute in host side)
- excute docker with following options: -v "$HOME/.Xauthority:/root/.Xauthority:rw" -v /tmp/.X11-unix:/tmp/.X11-unix -e DISPLAY=$DISPLAY

(0.74487168, 0.125)
(0.87307787, 0.25)
(0.89820999, 0.5)
(0.92245173, 1.0)
(0.93532807, 2.0)
(0.98055845, 4.0)
(0.99051809, 8.0)
0.906431
- https://luna16.grand-challenge.org/download/
- download data and candidates_V2.csv
- https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI
- download Radiologist Annotations/Segmentations (XML)
- https://drive.google.com/drive/u/1/folders/15d8BsRLNkroAl0iypHaIrlRorhBImzYk
- download detector.ckpt
- set luna and lidc path: set config_training.py
- export path: source export_path.sh
- train, val idx npy make: python preprocess/make_validate_npy.py
- preprocess data make: python preprocess/prepare.py
- train: sh train.sh
- make test result bbox: sh test.sh
- make froc sumbit: python make_FROC_submit_native.py
- calc froc curve: sh eval.sh
- apply UNET base lung segment
https://github.com/lfz/DSB2017
https://github.com/juliandewit/kaggle_ndsb2017