forked from migp11/PhysiBoSSv2-EMEWS
For results, please checkout to the 'coin_paper_results' branch
This repository combines various libaries and components to deliver a parallelized simulator for tumor cell growth that can be used for exploring parameter values of interest, for example related to cell growth, or to the effectiveness of drug treatments.
An EMEWS template is utilized, which is a workflow that incorporates Swift-T and enables the combination and parallel execution of model exploration algorithms written in different languages. Combined therein are:
- PhysiBoSSv2 with the spheroid-TNF-v2 model integrated as a custom module
- a Genetic Algorithm implementation based on the DEAP Python framework
- an Active Learning approach with Random Forests for parameter space characterization (as in Ozik et al., 2019)
- scripts for simulation results summarization
The spheroid-TNF-v2 is an implementation of a multi-scale agent-based model that simulates the growth of tumor spheroids. The model can take into account the effects of a signaling molecule that binds to cell receptors and may trigger a wide range of difference responses. Specifically, this workflow uses PhysiCell to provide the cell agents with an intracellular signal transduction model utilizing the PhysiBoSS extension. This signalling model is used to compute cell responses to perturbations such as the presence of signalling molecules and drugs.
This repository can be used to:
- Calibrate biophysical parameters of the given model based on some reference cases (golden standards)
- Find optimal treatments for reducing tumor cell population
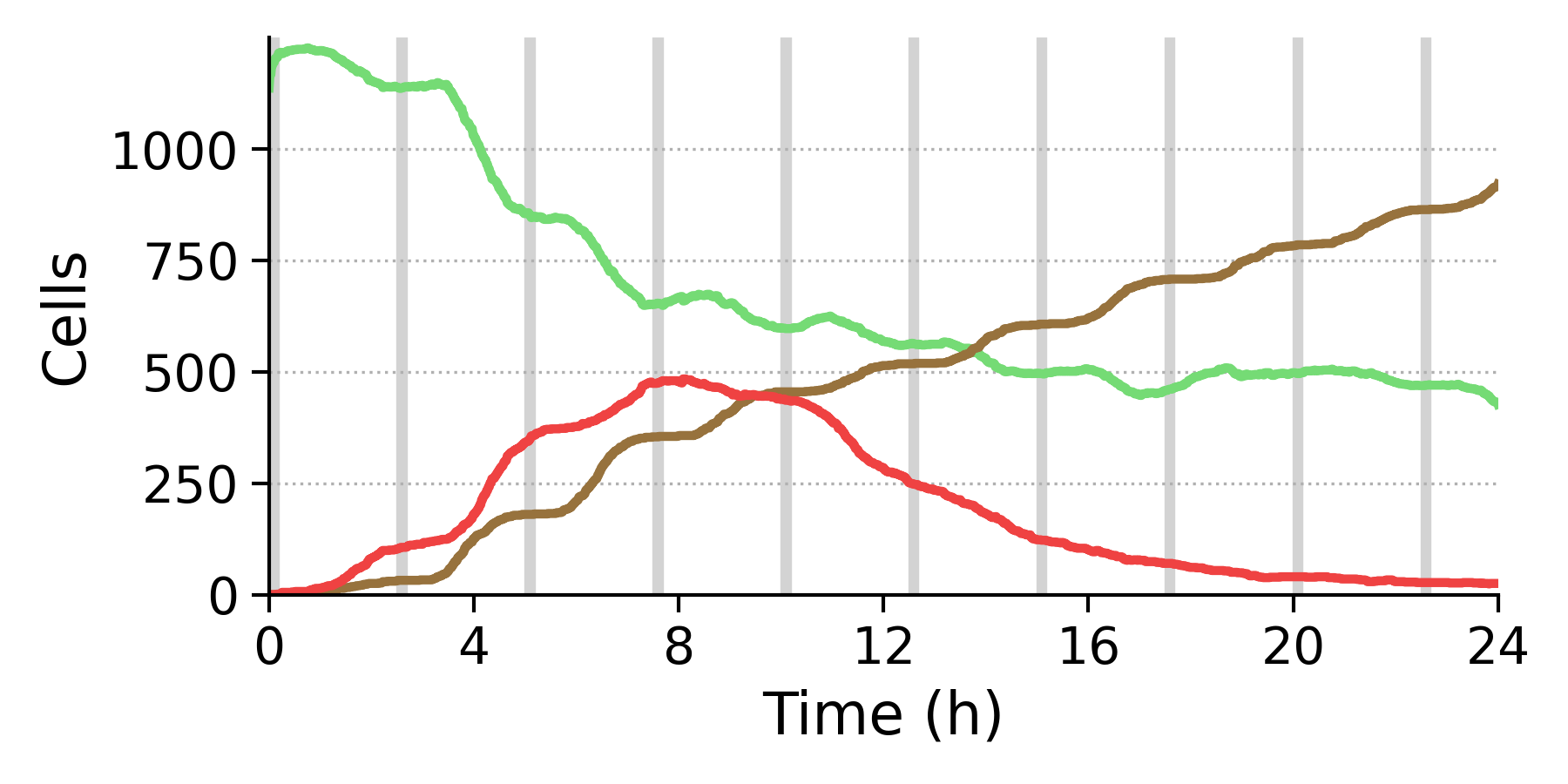
-Examines the differences in results between current simulator configurations and some 'Golden Standard' cases. Here, we have the following two.
Cell counts in the two Golden Standard cases (Green: Alive, Red: Apoptotic, Brown: Necrotic):
TNF frequency 150:
TNF frequency 600:
The parallel search is performed with the use of a Genetic Algorithm that has configurable objective functions as replaceable components.
-For calibrating the biophysical parameters, various distance metrics are utilized to measure the difference between the simulation output produced using the various candidate solutions, and the output of two ground truth simulations, which have predefined settings related to drug treatments and are produced by previous simulator versions. In particular, the distance types that can be used are the following:
- Euclidean distance
- Dynamic Time Warping
- l1 distance
and the parameters that are being explored are: TNFR Binding rate, TNFR Endocytosis rate, TNFR Recycling rate.
-For the evaluation of drug treatment configurations a different objective function is used, which counts the number of alive tumor cells at the end of the simulation, when applying treatments with the following configurable parameters: TNF Administration frequency, TNF Administration duration, TNF Concentration.
-The approach presented in Ozik et al., 2019, where Random Forests are used to classify the parameter space into interesting and not interesting regions.
Clone the repository:
$ git clone https://github.com/xarakas/spheroid-tnf-v2-emews.git
Compile [PhysiBoSSv2]:
$ cd data/PhysiBoSSv2
$ make
$ cd ../..
{
"distance_type" : "l1", // Can either be 'euclidean', 'dtw', or 'l1'
"termination_crit" : "fitmin", // Can either be 'genmax', 'fitmin', 'fitavg', or 'fitvar'
"termination_args" : 2.55, // Either integer or float, according to the "termination_crit" value (cf. below)
"pop_num" : 40, // Population number
"crossover_prob": 0.75, // Crossover probability
"mutation_prob": 0.5, // Mutation probability
"tournament_size": 3 // Tournament selection - tournament size
}
termination_crit:
genmax: Stop when a number of generations is met (the provided integer in the termination_args field).
fitmin: Stop when an individual with lower fitness than the termination_args value is discovered.
fitavg: Stop when the average fitness score of the population drops below termination_args value.
fitvar: Stop when the variance of the fitness scores of the population is below termination_args value for five consecutive generations.
The params file must be described in .json format, see examples in data/:
$ bash swift/swift_run_eqpy_compare.sh <EXPERIMENT_ID> <GA_PARAMS_FILE> <GA_CONFIG> <CHECKPOINT_FILE>
(e.g. bash swift/swift_run_eqpy_compare.sh experiment_1 data/ga_params.json data/ga_config.json last_experiment.pkl)
<CHECKPOINT_FILE> is optional, and contains the GA state from a previous run (stored automatically inside the experiment folder experiments/.
Logs regarding time, individuals examined and their fitness scores can be found in experiments/<EXPERIMENT_ID>/generations.log.
Again, a params file should be given, see e.g. data/inputs.txt:
$ bash swift/swift_run_sweep.sh <EXPERIMENT_ID> <SWEEP_INPUT>
(e.g. bash swift/swift_run_sweep.sh experiment_1 data/input.txt)
$ bash swift/swift_run_eqpy.sh <EXPERIMENT_ID> <GA_PARAMS_FILE> <GA_CONFIG> <CHECKPOINT_FILE>
(e.g. bash swift/swift_run_eqpy.sh experiment_1 data/ga_params.json data/ga_config.json last_experiment.pkl)
<CHECKPOINT_FILE> is optional, and contains the GA state from a previous run (stored automatically inside the experiment folder experiments/.
Logs regarding time, individuals examined and their fitness scores can be found in experiments/<EXPERIMENT_ID>/generations.log.
In the seeded scenario, in which a part of the initial population is set by the user, a txt file named interesting_points.txt containing the initial point must be provided in the python folder. If no such file is provided, then the initial population is created at random.
$ bash swift/swift_run_eqpy_sa.sh <EXPERIMENT_ID> <SA_PARAMS_FILE> <SA_CONFIG>
(e.g. bash swift/swift_run_eqpy_sa.sh experiment_1 data/ga_params.json data/sa_config.json)
The parameters of the simulated annealing method are set in the SA_CONFIG file, as follows:
{
"temperature" : 100, // Initial temperature
"min_temperature" : 15, // Minimum temperature
"cooling" : 0.8, // Cooling factor
"max_iterations" : 10, // Iterations per temperature level
"distance_threshold" : 0.02, // Distance threshold of neighbourhood
"seeded_pop": "YES" // 'YES' if initial point is given from txt file
}
If seeded_pop is set to "YES", a txt file named best_point.txt containing the initial point must be provided in the python folder.
$ bash swift/swift_run_eqpy_rand.sh <EXPERIMENT_ID> <RAND_PARAMS_FILE> <RAND_CONFIG>
(e.g. bash swift/swift_run_eqpy_rand.sh experiment_1 data/ga_params.json data/rand_config.json)
Logs regarding the experiment conducted can be found in experiments/<EXPERIMENT_ID>/iterations.log.
Scripts regarding the analysis of the experiments and the graphical representation of the various parts of the experiments can be found in the folder experimental_analysis.
The parameters of the active learning algorithm are set in the RAND_CONFIG file, as follows:
{
"method" : "BIRCH", // Can either be 'BIRCH', 'DBSCAN', or 'KMEANS'
"param1" : 100, // KMEANS->k, DBSCAN->eps, BIRCH->branching factor
"param2" : 0.1, // KMEANS->None, DBSCAN->MinPts, BIRCH->threshold
"prev_results": "YES" // 'YES' if results from past experiments are used
}
The random seed of the experiment conducted and the number of the initial population is set in the swift/swift_run_eqpy_rand.sh file.
Results from past experiments can used in order to reduce the runtime of the active learning scenario. These results should be provided with a csv file named all_exps_DD.csv csv file in the python folder.
.sh files in the swift/ folder set various environment parameters for the execution.
Make sure that:
- PROCS is set to a number higher than 3.
- MACHINE is set to blank ("") if not having
slurminstalled (e.g. when executing locally and not on HPC)
This project is compatible with swift-t v. 1.3+. Earlier versions will NOT work.
For more information regarding installation and execution, please see the file how-to-launch.txt