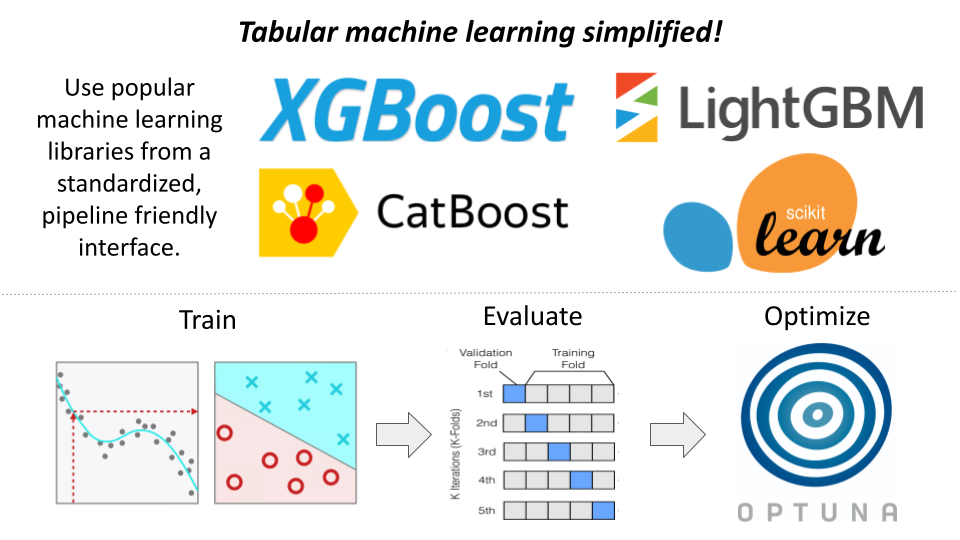
I've packaged and open sourced my personal machine learning tools to speed up your next data science project.
Train, evaluate, ensemble, and optimize hyperparameters from a standardized interface.
- Train models efficiently without worrying about library differences!
tabular_mlimplements library specific, performance oriented, patterns/classes under-the-hood (i.e.,xgboost.DMatrix -> xgboost.Booster). - Automate the K-Fold evaluation process across multiple models simultaneously (including ensembles).
- Rapidly optimize hyperparameters using
optuna. Leverage our built-in parameter search spaces, or adjust to your needs. - Plugin-able. Write your own plugins to extend functionality without forking (and consider contributing your plugins!).
Contents:
This library is available on PyPI and can be easily pip installed into your environment.
pip install tabular-ml
This library wraps each regression and classification model in a type= tabular_ml.MLModel object. This allows models to be used interchangeably without worrying about library specific nuance.
We use tabular_ml.ModelFactory to keep track of all supported models. One can programmatically explore model offerings with the following functions:
import tabular_ml
# to get a list of regression models
tabular_ml.ModelFactory.get_regression_models()
# to get a list of classification models
tabular_ml.ModelFactory.get_regression_models()
# get a dictionary storing both the above lists
tabular_ml.ModelFactory.get_all_models()It is also best practice to get model_objects from the factory rather than importing from the tabular_ml.ml_models module. Below we demonstrate getting the CatBoostRegressionModel object.
import tabular_ml
# get the model from the factory
obj = tabular_ml.ModelFactory.get_model('CatBoostRegressionModel')
# use it!
trained_model = obj.train_model(
params={'iterations': 500}...
)CatBoostRegressionModelCatBoostClassificationModel
XGBoostRegressionModelXGBoostClassificationModel
LightGBMRegressionModelLightGBMClassificationModel
LinearRegressionModelRidgeRegressionModelLassoRegressionModelElasticNetRegressionModelBayesianRidgeRegressionModel
Each model registered to ModelFactory must be a concrete implementation of the tabular_ml.base.MLModel abstract base class.
This means that all registered models contain the following functions allowing them to be used interchangeably:
def train_model(
x_train: pd.DataFrame,
y_train: pd.Series,
model_params: Dict[str, Any],
weights_train: Optional[pd.Series] = None,
categorical_features: Optional[List[str]] = None,
) -> object: # type depends on the specific model implementation
"""Train a model instance.
Arguments:
x_train: training data features.
y_train: training data target.
model_params: dictionary of model parameters.
weights_train: optional training data weights.
categorical_features: optional list of categorical features.
Returns:
trained model instance. (i.e. xgboost.Booster).
"""
...
def make_predictions(
trained_model: object, # same as train_model() output!
x_test: pd.DataFrame,
categorical_features: Optional[List[str]] = None,
) -> np.ndarray:
"""Make predictions with a trained model instance.
Arguments:
trained_model: trained model instance.
x_test: test data features.
categorical_features: optional list of categorical features.
Returns:
Predictions as a numpy array.
"""
...
def train_and_predict(
x_train: pd.DataFrame,
y_train: pd.Series,
x_test: pd.DataFrame,
model_params: Dict[str, Any],
weights_train: Optional[pd.Series] = None,
categorical_features: Optional[List[str]] = None,
) -> Tuple[object, np.ndarray]:
"""Trains a model and makes predictions on param:x_test.
Wraps train_model() and make_predictions() into a single function.
Arguments:
x_train: training data features.
y_train: training data target.
x_test: test data features.
model_params: dictionary of model parameters.
weights_train: optional training data weights.
categorical_features: optional list of categorical features.
Returns:
Trained model instance and predictions.
"""
...
def objective(
trial: Trial,
features: pd.DataFrame,
target: pd.Series,
kfolds: int,
metric_function: callable,
weights: Optional[pd.Series] = None,
categorical_features: Optional[List[str]] = None,
random_state: Optional[int] = None,
custom_param_ranges: Optional[OptunaRangeDict] = None,
) -> float:
"""Controls optuna hyperparameter optimization behavior.
NOTE: This function is not intended to be used directly. It is called
by tabular_ml.find_optimal_parameters().
Arguments:
trial: optuna.Trial object.
features: training data features.
target: training data target.
kfolds: number of folds to use for cross validation.
metric_function: function to use for evaluation.
weights: optional training data weights.
categorical_features: optional list of categorical features.
random_state: optional random state.
custom_param_ranges: optional dictionary of custom parameter ranges.
Ex: {'param_name': (min_val, max_val)}.
Returns:
Metric value to minimize after running K-fold evaluation.
"""
...To extend the functionality of this library one can simply "register" your custom MLModel implementation to ModelFactory. This is demonstrated below. Note that all MLModel functions and expended class variables must be present. Functions can remain empty if necessary.
import tabular_ml
# write a custom MLModel to import
class CustomRegressionModel(tabular_ml.MLModel):
model_type = 'regression'
def train_model():
...
def make_predictions():
...
def train_and_predict():
...
def optimize():
...
# register your MLModel to the factory!
ModelFactory.register_model(CustomRegressionModel)That said we highly encourage making a pull request and contributing your custom MLModel such that it can be enjoyed by the world.
To evaluate a given model or set of models using K-Fold Cross Validation one can use the tabular_ml.k_fold_cv() function. This function is designed to be flexible and can be used to evaluate any set of models simultaneously by simply passing a list of model names and a dictionary of model parameters into param:model_names and param:model_params respectively.
Key points:
- The names past in should match the offerings from
ModelFactory.get_all_models(). - To control the # of folds one can pass an integer into param:
n_splits. By default this is set to 5. - Results are stored in a
KFoldOutputdataclass. This dataclass contains all relevant results, trained model instances for further inspection, run-time, and more. - When multiple model names are passed in results are stored for each model individually as well as all the ensemble average of all model predictions.
- Any metric function can be used to evaluate the models. By default we use R-Squared for regression and AUC for classification. To use a custom metric function simply pass it into param:
metric_function. The function should have the signaturef(y_true, y_preds, **kwargs) -> float.
The function documentation is provided below:
def k_fold_cv(
x_data: pd.DataFrame,
y_data: pd.Series,
model_names: List[str],
model_params: Dict[str, Dict[str, float | int | str]],
weights_data: Optional[pd.Series] = None,
categorical_features: Optional[List[str]] = None,
n_splits: Optional[int] = None,
metric_function: Optional[callable] = None,
metric_function_kwargs: Optional[Dict[str, Any]] = None,
random_state: Optional[int] = None,
logging_file_path: Optional[str | Path] = None,
) -> KFoldOutput:
"""Runs K-fold CV for any set of models.
Arguments:
x_data: A pandas DataFrame of features.
y_data: A pandas Series of targets.
model_names: A list of valid model names.
Valid names can be queried using ModelFactory.get_all_models().
model_params: a dict with model names as keys,
and parameters for that model as values.
weights_data: A pandas Series of training weights.
categorical_features: A list of categorical feature names.
n_splits: # of K-Folds. Default is 5.
metric_function: A function with the signature
f(y_true, y_preds, **kwargs) -> float. Default is R-Squared.
metric_function_kwargs: Kwargs to pass to the metric function.
random_state: Random state to use for K-Folds.
logging_file_path: Path to a log file.
Returns:
A populated KFoldOutput dataclass with relevant info.
"""Optuna is an open-source hyperparameter optimization framework. This library provides a wrapper around optuna to make hyperparameter optimization as simple as calling tabular_ml.find_optimal_parameters() and passing in the necessary arguments. Under-the-hood, this function uses K-Fold CV to evaluate each trial in relation to a user-provided "metric function" (i.e., sklearn.metrics.mae).
Key Points:
- Pass in a model to optimize by setting param:
modelto it's factory registered name (i.e., "CatBoostRegressionModel"). - A metric function must be passed in to evaluate the model! By default we use R-Squared for regression and AUC for classification. To use a custom metric function simply pass it into param:
metric_function. The function should have the signaturef(y_true, y_preds, **kwargs) -> float. - Each model has a default set of parameters to optimize, and range of values to. To override these defaults pass in a dictionary of custom ranges into param:
custom_optuna_ranges. The dictionary should have parameter names as keys and a tuple of(min_val, max_val)as values. For example, to optimizen_estimatorsforXGBoostRegressionModelone could pass in{'n_estimators': (100, 1000)}.
This function's arguments are shown below:
def find_optimal_parameters(
model: str,
features: pd.DataFrame,
target: pd.Series,
metric_function: callable,
direction: Optional[str] = None,
n_trials: Optional[int] = None,
timeout: Optional[int] = None,
kfolds: Optional[int] = None,
weights: Optional[pd.Series] = None,
categorical_features: Optional[List[str]] = None,
random_state: Optional[int] = None,
custom_optuna_ranges: Optional[OptunaRangeDict] = None,
logging_file_path: Optional[str | Path] = None,
) -> Dict[str, Any]:
"""Runs optuna optimization for a MLModel.
Arguments:
model: A valid model name.
Valid names can be queried using ModelFactory.get_all_models().
features: A pandas DataFrame of features.
target: A pandas Series of targets.
metric_function: A function with the signature
f(y_true, y_preds, **kwargs) -> float.
direction: 'maximize' or 'minimize' the metric function.
If None, will infer direction from metric function.
n_trials: # of trials to run. Default is 20.
timeout: # of seconds to run.
kfolds: # of K-Folds. Default is 5.
weights: A pandas Series of training weights.
categorical_features: A list of categorical feature names.
random_state: Random state to use for K-Folds.
custom_optuna_ranges: A dict of custom optuna ranges.
logging_file_path: Path to a log file.
Returns:
A dict of optimal parameters for the model.
"""import tabular_ml
import sklearn.metrics
# see model offerings
print(tabular_ml.ModelFactory.get_regression_models())
# get a model object
xgboost_model = tabular_ml.ModelFactory.get_model('XGBoostRegressionModel')
# pull in data
data_df = pd.read_csv('data.csv')
# select parameters
model_params = {
'n_estimators': 100,
'max_depth': 3,
'learning_rate': 0.1,
'objective': 'reg:squarederror',
}
# evaluate the model with 5-Fold CV (using MAE)
results = tabular_ml.k_fold_cv(
x_data=data_df.drop(columns=['target']),
y_data=data_df['target'],
model_names=['XGBoostRegressionModel'],
model_params={'XGBoostRegressionModel': model_params},
n_splits=5,
metric_function=sklearn.metrics.mae,
random_state=5,
)
# print results dataclass
print(results)import tabular_ml
import sklearn.metrics
# see model offerings
print(tabular_ml.ModelFactory.get_classification_models())
# optimize a model to minimize log-loss over 25 trials
optimal_params = tabular_ml.find_optimal_parameters(
x_data=data_df.drop(columns=['target']),
y_data=data_df['target'],
model_name='LightGBMClassificationModel',
n_trials=25,
n_splits=5,
metric_function=sklearn.metrics.log_loss,
)
# train a final model with the optimal params
model = tabular_ml.ModelFactory.get_model('LightGBMClassificationModel')
trained_model = model.train_model(
x_train=data_df.drop(columns=['target']),
y_train=data_df['target'],
model_params=optimal_params,
# make predictions with the trained model
preds = model.make_predictions(
trained_model=trained_model,
x_test=pd.read_csv('test_data.csv'),
)Contributions are welcome!
The easiest contribution to make is to add your own MLModel implementation. This can be done by simply extending the tabular_ml.base.MLModel abstract base class and registering your model to ModelFactory. To do this simply decorate your class with @ModelFactory.implemented_model. This allows the factory to register the model on import.
Regardless of what one contributes, before making a pull request please run the following commands to ensure your code is formatted correctly and passes all tests.
(base) cd Repo/Path/Tabular_Ml
# install our dev environment
(base) conda env create -f environment.yml
(base) conda activate tabular_ml
# run pre-commit for code formatting
(tabular_ml) pre-commit run --all-files
# run our test suite
(tabular_ml) pytest
The pre-commit may have to be ran multiple times if many problems are found. Keep repeating the command until all checks pass.