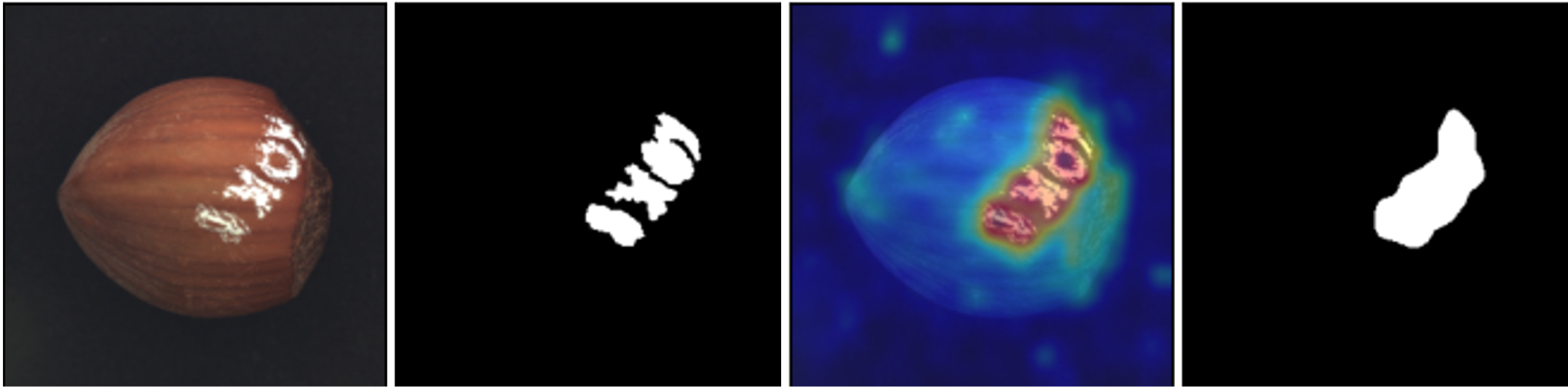
A library for benchmarking, developing and deploying deep learning anomaly detection algorithms
Anomalib is a deep learning library that aims to collect state-of-the-art anomaly detection algorithms for benchmarking on both public and private datasets. Anomalib provides several ready-to-use implementations of anomaly detection algorithms described in the recent literature, as well as a set of tools that facilitate the development and implementation of custom models. The library has a strong focus on image-based anomaly detection, where the goal of the algorithm is to identify anomalous images, or anomalous pixel regions within images in a dataset. Anomalib is constantly updated with new algorithms and training/inference extensions, so keep checking!
Key features:
- The largest public collection of ready-to-use deep learning anomaly detection algorithms and benchmark datasets.
- PyTorch Lightning based model implementations to reduce boilerplate code and limit the implementation efforts to the bare essentials.
- All models can be exported to OpenVINO Intermediate Representation (IR) for accelerated inference on intel hardware.
- A set of inference tools for quick and easy deployment of the standard or custom anomaly detection models.
To get an overview of all the devices where anomalib as been tested thoroughly, look at the Supported Hardware section in the documentation.
You can get started with anomalib by just using pip.
pip install anomalibIt is highly recommended to use virtual environment when installing anomalib. For instance, with anaconda, anomalib could be installed as,
yes | conda create -n anomalib_env python=3.8
conda activate anomalib_env
git clone https://github.com/openvinotoolkit/anomalib.git
cd anomalib
pip install -e .By default python tools/train.py
runs PADIM model on leather category from the MVTec AD (CC BY-NC-SA 4.0) dataset.
python tools/train.py # Train PADIM on MVTec AD leatherTraining a model on a specific dataset and category requires further configuration. Each model has its own configuration
file, config.yaml
, which contains data, model and training configurable parameters. To train a specific model on a specific dataset and
category, the config file is to be provided:
python tools/train.py --config <path/to/model/config.yaml>For example, to train PADIM you can use
python tools/train.py --config anomalib/models/padim/config.yamlNote that --model_config_path will be deprecated in v0.2.8 and removed
in v0.2.9.
Alternatively, a model name could also be provided as an argument, where the scripts automatically finds the corresponding config file.
python tools/train.py --model padimwhere the currently available models are:
It is also possible to train on a custom folder dataset. To do so, data section in config.yaml is to be modified as follows:
dataset:
name: <name-of-the-dataset>
format: folder
path: <path/to/folder/dataset>
normal: normal # name of the folder containing normal images.
abnormal: abnormal # name of the folder containing abnormal images.
task: segmentation # classification or segmentation
mask: <path/to/mask/annotations> #optional
extensions: null
split_ratio: 0.2 # ratio of the normal images that will be used to create a test split
seed: 0
image_size: 256
train_batch_size: 32
test_batch_size: 32
num_workers: 8
transform_config: null
create_validation_set: true
tiling:
apply: false
tile_size: null
stride: null
remove_border_count: 0
use_random_tiling: False
random_tile_count: 16Anomalib contains several tools that can be used to perform inference with a trained model. The script in tools/inference contains an example of how the inference tools can be used to generate a prediction for an input image.
If the specified weight path points to a PyTorch Lightning checkpoint file (.ckpt), inference will run in PyTorch. If the path points to an ONNX graph (.onnx) or OpenVINO IR (.bin or .xml), inference will run in OpenVINO.
The following command can be used to run inference from the command line:
python tools/inference.py \
--config <path/to/model/config.yaml> \
--weight_path <path/to/weight/file> \
--image_path <path/to/image>As a quick example:
python tools/inference.py \
--config anomalib/models/padim/config.yaml \
--weight_path results/padim/mvtec/bottle/weights/model.ckpt \
--image_path datasets/MVTec/bottle/test/broken_large/000.pngIf you want to run OpenVINO model, ensure that openvino apply is set to True in the respective model config.yaml.
optimization:
openvino:
apply: trueExample OpenVINO Inference:
python tools/inference.py \
--config \
anomalib/models/padim/config.yaml \
--weight_path \
results/padim/mvtec/bottle/compressed/compressed_model.xml \
--image_path \
datasets/MVTec/bottle/test/broken_large/000.png \
--meta_data \
results/padim/mvtec/bottle/compressed/meta_data.jsonEnsure that you provide path to
meta_data.jsonif you want the normalization to be applied correctly.
anomalib supports MVTec AD (CC BY-NC-SA 4.0) and BeanTech (CC-BY-SA) for benchmarking and folder for custom dataset training/inference.
MVTec AD dataset is one of the main benchmarks for anomaly detection, and is released under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License (CC BY-NC-SA 4.0).
| Model | Avg | Carpet | Grid | Leather | Tile | Wood | Bottle | Cable | Capsule | Hazelnut | Metal Nut | Pill | Screw | Toothbrush | Transistor | Zipper | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PatchCore | Wide ResNet-50 | 0.980 | 0.984 | 0.959 | 1.000 | 1.000 | 0.989 | 1.000 | 0.990 | 0.982 | 1.000 | 0.994 | 0.924 | 0.960 | 0.933 | 1.000 | 0.982 |
| PatchCore | ResNet-18 | 0.973 | 0.970 | 0.947 | 1.000 | 0.997 | 0.997 | 1.000 | 0.986 | 0.965 | 1.000 | 0.991 | 0.916 | 0.943 | 0.931 | 0.996 | 0.953 |
| CFlow | Wide ResNet-50 | 0.962 | 0.986 | 0.962 | 1.0 | 0.999 | 0.993 | 1.0 | 0.893 | 0.945 | 1.0 | 0.995 | 0.924 | 0.908 | 0.897 | 0.943 | 0.984 |
| PaDiM | Wide ResNet-50 | 0.950 | 0.995 | 0.942 | 1.0 | 0.974 | 0.993 | 0.999 | 0.878 | 0.927 | 0.964 | 0.989 | 0.939 | 0.845 | 0.942 | 0.976 | 0.882 |
| PaDiM | ResNet-18 | 0.891 | 0.945 | 0.857 | 0.982 | 0.950 | 0.976 | 0.994 | 0.844 | 0.901 | 0.750 | 0.961 | 0.863 | 0.759 | 0.889 | 0.920 | 0.780 |
| STFPM | Wide ResNet-50 | 0.876 | 0.957 | 0.977 | 0.981 | 0.976 | 0.939 | 0.987 | 0.878 | 0.732 | 0.995 | 0.973 | 0.652 | 0.825 | 0.5 | 0.875 | 0.899 |
| STFPM | ResNet-18 | 0.893 | 0.954 | 0.982 | 0.989 | 0.949 | 0.961 | 0.979 | 0.838 | 0.759 | 0.999 | 0.956 | 0.705 | 0.835 | 0.997 | 0.853 | 0.645 |
| DFM | Wide ResNet-50 | 0.891 | 0.978 | 0.540 | 0.979 | 0.977 | 0.974 | 0.990 | 0.891 | 0.931 | 0.947 | 0.839 | 0.809 | 0.700 | 0.911 | 0.915 | 0.981 |
| DFM | ResNet-18 | 0.894 | 0.864 | 0.558 | 0.945 | 0.984 | 0.946 | 0.994 | 0.913 | 0.871 | 0.979 | 0.941 | 0.838 | 0.761 | 0.95 | 0.911 | 0.949 |
| DFKDE | Wide ResNet-50 | 0.774 | 0.708 | 0.422 | 0.905 | 0.959 | 0.903 | 0.936 | 0.746 | 0.853 | 0.736 | 0.687 | 0.749 | 0.574 | 0.697 | 0.843 | 0.892 |
| DFKDE | ResNet-18 | 0.762 | 0.646 | 0.577 | 0.669 | 0.965 | 0.863 | 0.951 | 0.751 | 0.698 | 0.806 | 0.729 | 0.607 | 0.694 | 0.767 | 0.839 | 0.866 |
| GANomaly | 0.421 | 0.203 | 0.404 | 0.413 | 0.408 | 0.744 | 0.251 | 0.457 | 0.682 | 0.537 | 0.270 | 0.472 | 0.231 | 0.372 | 0.440 | 0.434 |
| Model | Avg | Carpet | Grid | Leather | Tile | Wood | Bottle | Cable | Capsule | Hazelnut | Metal Nut | Pill | Screw | Toothbrush | Transistor | Zipper | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PatchCore | Wide ResNet-50 | 0.980 | 0.988 | 0.968 | 0.991 | 0.961 | 0.934 | 0.984 | 0.988 | 0.988 | 0.987 | 0.989 | 0.980 | 0.989 | 0.988 | 0.981 | 0.983 |
| PatchCore | ResNet-18 | 0.976 | 0.986 | 0.955 | 0.990 | 0.943 | 0.933 | 0.981 | 0.984 | 0.986 | 0.986 | 0.986 | 0.974 | 0.991 | 0.988 | 0.974 | 0.983 |
| CFlow | Wide ResNet-50 | 0.971 | 0.986 | 0.968 | 0.993 | 0.968 | 0.924 | 0.981 | 0.955 | 0.988 | 0.990 | 0.982 | 0.983 | 0.979 | 0.985 | 0.897 | 0.980 |
| PaDiM | Wide ResNet-50 | 0.979 | 0.991 | 0.970 | 0.993 | 0.955 | 0.957 | 0.985 | 0.970 | 0.988 | 0.985 | 0.982 | 0.966 | 0.988 | 0.991 | 0.976 | 0.986 |
| PaDiM | ResNet-18 | 0.968 | 0.984 | 0.918 | 0.994 | 0.934 | 0.947 | 0.983 | 0.965 | 0.984 | 0.978 | 0.970 | 0.957 | 0.978 | 0.988 | 0.968 | 0.979 |
| STFPM | Wide ResNet-50 | 0.903 | 0.987 | 0.989 | 0.980 | 0.966 | 0.956 | 0.966 | 0.913 | 0.956 | 0.974 | 0.961 | 0.946 | 0.988 | 0.178 | 0.807 | 0.980 |
| STFPM | ResNet-18 | 0.951 | 0.986 | 0.988 | 0.991 | 0.946 | 0.949 | 0.971 | 0.898 | 0.962 | 0.981 | 0.942 | 0.878 | 0.983 | 0.983 | 0.838 | 0.972 |
| Model | Avg | Carpet | Grid | Leather | Tile | Wood | Bottle | Cable | Capsule | Hazelnut | Metal Nut | Pill | Screw | Toothbrush | Transistor | Zipper | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PatchCore | Wide ResNet-50 | 0.976 | 0.971 | 0.974 | 1.000 | 1.000 | 0.967 | 1.000 | 0.968 | 0.982 | 1.000 | 0.984 | 0.940 | 0.943 | 0.938 | 1.000 | 0.979 |
| PatchCore | ResNet-18 | 0.970 | 0.949 | 0.946 | 1.000 | 0.98 | 0.992 | 1.000 | 0.978 | 0.969 | 1.000 | 0.989 | 0.940 | 0.932 | 0.935 | 0.974 | 0.967 |
| CFlow | Wide ResNet-50 | 0.944 | 0.972 | 0.932 | 1.0 | 0.988 | 0.967 | 1.0 | 0.832 | 0.939 | 1.0 | 0.979 | 0.924 | 0.971 | 0.870 | 0.818 | 0.967 |
| PaDiM | Wide ResNet-50 | 0.951 | 0.989 | 0.930 | 1.0 | 0.960 | 0.983 | 0.992 | 0.856 | 0.982 | 0.937 | 0.978 | 0.946 | 0.895 | 0.952 | 0.914 | 0.947 |
| PaDiM | ResNet-18 | 0.916 | 0.930 | 0.893 | 0.984 | 0.934 | 0.952 | 0.976 | 0.858 | 0.960 | 0.836 | 0.974 | 0.932 | 0.879 | 0.923 | 0.796 | 0.915 |
| STFPM | Wide ResNet-50 | 0.926 | 0.973 | 0.973 | 0.974 | 0.965 | 0.929 | 0.976 | 0.853 | 0.920 | 0.972 | 0.974 | 0.922 | 0.884 | 0.833 | 0.815 | 0.931 |
| STFPM | ResNet-18 | 0.932 | 0.961 | 0.982 | 0.989 | 0.930 | 0.951 | 0.984 | 0.819 | 0.918 | 0.993 | 0.973 | 0.918 | 0.887 | 0.984 | 0.790 | 0.908 |
| DFM | Wide ResNet-50 | 0.918 | 0.960 | 0.844 | 0.990 | 0.970 | 0.959 | 0.976 | 0.848 | 0.944 | 0.913 | 0.912 | 0.919 | 0.859 | 0.893 | 0.815 | 0.961 |
| DFM | ResNet-18 | 0.919 | 0.895 | 0.844 | 0.926 | 0.971 | 0.948 | 0.977 | 0.874 | 0.935 | 0.957 | 0.958 | 0.921 | 0.874 | 0.933 | 0.833 | 0.943 |
| DFKDE | Wide ResNet-50 | 0.875 | 0.907 | 0.844 | 0.905 | 0.945 | 0.914 | 0.946 | 0.790 | 0.914 | 0.817 | 0.894 | 0.922 | 0.855 | 0.845 | 0.722 | 0.910 |
| DFKDE | ResNet-18 | 0.872 | 0.864 | 0.844 | 0.854 | 0.960 | 0.898 | 0.942 | 0.793 | 0.908 | 0.827 | 0.894 | 0.916 | 0.859 | 0.853 | 0.756 | 0.916 |
| GANomaly | 0.834 | 0.864 | 0.844 | 0.852 | 0.836 | 0.863 | 0.863 | 0.760 | 0.905 | 0.777 | 0.894 | 0.916 | 0.853 | 0.833 | 0.571 | 0.881 |
If you use this library and love it, use this to cite it 🤗
@misc{anomalib,
title={Anomalib: A Deep Learning Library for Anomaly Detection},
author={Samet Akcay and
Dick Ameln and
Ashwin Vaidya and
Barath Lakshmanan and
Nilesh Ahuja and
Utku Genc},
year={2022},
eprint={2202.08341},
archivePrefix={arXiv},
primaryClass={cs.CV}
}