Requires prior basic knowledge about Tract-Based Spatial Statistics. Requires nilearn and nipype.
See this post for a few explanations on why and how.
The following splits a .con contrast file into multiple ones and generates a series of independent commands to run randomise on the new contrast files in parallel jobs.
from tbss import contrasts
commands = contrasts.randomise_parallel(in_file = '/path/to/all_FA_skeletonised.nii.gz',
out_basename = '/path/to/tbss_FA',
mask = '/path/to/mean_FA_skeleton_mask.nii.gz',
tcon = '/path/to/contrasts.con',
design_mat = '/path/to/design.mat',
num_perm = 1000,
n_cpus = 6,
email_notify = None,
sleep_interval = 120)Temporary contrast files: ['/path/to/tbss_FA_part0.con', '/path/to/tbss_FA_part1.con', '/path/to/tbss_FA_part2.con', '/path/to/tbss_FA_part3.con', '/path/to/tbss_FA_part4.con', '/path/to/tbss_FA_part5.con']
[u'sleep 0 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part0.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=1 -V &> /tmp/tmpHj9gPW.log',
u'sleep 120 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part1.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=2 -V &> /tmp/tmpDP2rM9.log',
u'sleep 240 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part2.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=3 -V &> /tmp/tmp5u74y0.log',
u'sleep 360 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part3.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=4 -V &> /tmp/tmpfIf08c.log',
u'sleep 480 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part4.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=5 -V &> /tmp/tmp0fhKN5.log',
u'sleep 600 ; randomise -i /path/to/all_FA_skeletonised.nii.gz -d /path/to/design.mat -t /path/to/tbss_FA_part5.con -m /path/to/mean_FA_skeleton_mask.nii.gz -n 1000 -R --T2 -o /path/to/tbss_FA --skipTo=6 -V &> /tmp/tmpPaWk5L.log']
n_cpussets the number of available CPU cores thatrandomisewill run on simultaneously (consequently the number of.confiles the contrasts will be split into) (default: 6)- If
email_notifyis an email address, notifications will be sent to that address before and after every job (default: None) sleep_intervalsets the interval in seconds before running the following job (will avoid RAM exhaustion whenrandomisestarts loading the data)
There are a few hard-coded options in the command call such as --T2 or -R. These can be easily modified in the code if necessary.
The produced commands can finally be stored in a shell script e.g. that may be passed to GNU parallel for execution, as in the following example:
cat /tmp/script.sh | parallel -j <n_cpus>
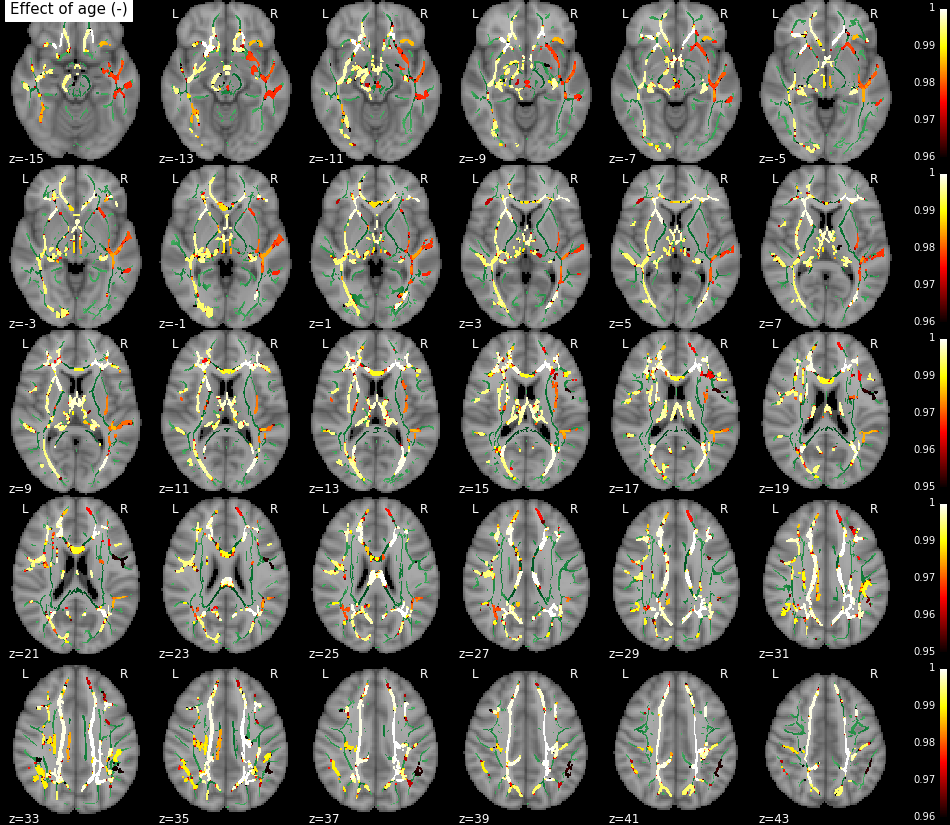
Requires nilearn.
from tbss import plotting
plotting.plot_stat_map('/path/to/tbss_tstat1.nii.gz',
'/path/to/mean_FA_skeleton.nii.gz',
start=-15,
end=43,
row_l=6,
step=2,
title='Effect of age (-)')