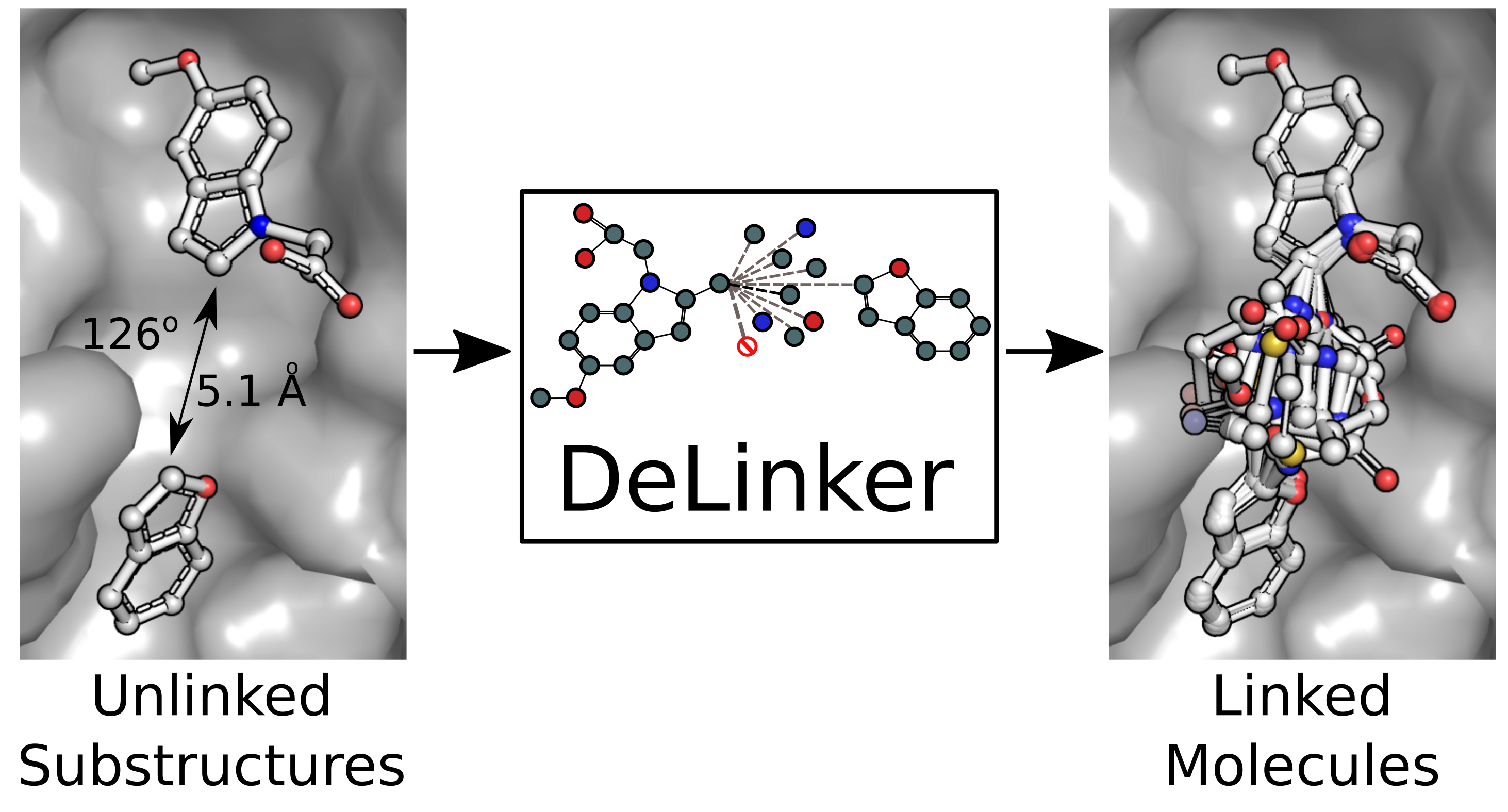
DeLinker - Deep Generative Models for 3D Linker Design
This repository contains our implementation of Deep Generative Models for 3D Linker Design (DeLinker).
If you found DeLinker useful, please cite our paper:
Imrie F, Bradley AR, van der Schaar M, Deane CM. Deep Generative Models for 3D Linker Design. Journal of Chemical Information and Modeling. 2020
@Article{Imrie2020,
author={Imrie, Fergus and Bradley, Anthony R. and van der Schaar, Mihaela and Deane, Charlotte M.},
title={Deep Generative Models for 3D Linker Design},
journal={Journal of Chemical Information and Modeling},
year={2020},
month={Mar},
day={20},
publisher={American Chemical Society},
issn={1549-9596},
doi={10.1021/acs.jcim.9b01120},
url={https://doi.org/10.1021/acs.jcim.9b01120}
}
Requirements
This code was tested in Python 3.6 with Tensorflow 1.10.
A yaml file containing all requirements is provided. This can be readily setup using conda.
conda env create -f DeLinker-env.yml
conda activate DeLinker-env
Data Extraction
Two primary datasets (ZINC and CASF) are in use.
To preprocess these datasets, please go to data directory and run prepare_data.py.
python prepare_data.py
Running DeLinker
We provide two settings of DeLinker. The first setting generates linkers with the same number of atoms as the reference molecule. The second setting generates linkers with a specified number of atoms.
To train and generate molecules using the first setting, use:
python DeLinker.py --dataset zinc --config '{"num_epochs": 10, "epoch_to_generate": 10, "train_file": "data/molecules_zinc_train.json", "valid_file": "data/molecules_zinc_valid.json"}'
To generate molecules with a pretrained model using the first setting, use
python DeLinker.py --dataset zinc --restore models/pretrained_DeLinker_model.pickle --config '{"generation": true, "number_of_generation_per_valid": 250, "batch_size": 1, "train_file": "data/molecules_zinc_test.json", "valid_file": "data/molecules_zinc_test.json"}'
To generate molecules using the second setting, use
python DeLinker_test.py --dataset zinc --restore models/pretrained_DeLinker_model.pickle --config '{"generation": true, "number_of_generation_per_valid": 250, "batch_size": 1, "train_file": "data/molecules_zinc_test_mode2.json", "valid_file": "data/molecules_zinc_test_mode2.json", "min_atoms": 3, "max_atoms": 11}'
In both cases, the output is of the following format:
Input fragments (SMILES) Ground truth molecule/fragments (SMILES) Generated molecule (SMILES)
More configurations can be found at function default_params in DeLinker.py.
Evaluation
A script to evaluate the generated molecules is provided in analysis directory.
python evaluate_generated_mols.py ZINC|CASF PATH_TO_GENERATED_MOLS PATH_TO_REFERENCE_MOLS ../data/data_zinc_final_train.txt SAVE_PATH OUTPUT_NAME NUM_CORES True None ./wehi_pains.csv >> log.txt
Pretrained Models and Generated Molecules
We provide a pretrained model:
models/pretrained_DeLinker_model.pickle
Generated molecules can be obtained upon request.
Examples
An example Jupyter notbook demonstrating the use of DeLinker for fragment linking can be found in the examples directory.
Contact (Questions/Bugs/Requests)
Please submit a Github issue or contact Fergus Imrie imrie@stats.ox.ac.uk.