gif provided by the awesome Jen Bright @MorphobeakGeek!
In this exercise we will use a github repo to collaboratively collate and simulate evolutionary trajectories for each participants' species body size using a simple brownian motion evolutionary model. This assumes evolutionary steps to progress comletely at random. You could say, it's a bit of lottery!
- Each participant will create and contribute a file specifying the parameters required to simulate and plot their species evolutionary trajectory.
- We'll collect all participants' files in the master repo.
- Once all trajectories are simulated they'll be plotted together.
- Participants will then get to see the skull and beak shape corresponding to their species relative body size!
NHM STARS training course: Thursday, 18th May 11:00-12:00
We'll be accepting pull requests by remote participants between 11.30 - 12.00, so anyone can get involved! Follow #EvoLottery on the day for live updates on twitter.
- fork the repo into your own account
- copy repo url link local files to github repo
- create new project
- checkout from version control/git
- paste github repo link
A template is provided in the repo, in folder /params named params_tmpl.R. Open the params_tmpl.R and save as to create a duplicate template. Name the template using the name of your species.
The parameters each participants need to supply are:
-
sig2: A numeric value greater than 0 but smaller than 5 -
species.name: a character string e.g."anas_krystallinus". Try to create a species name out of your name! -
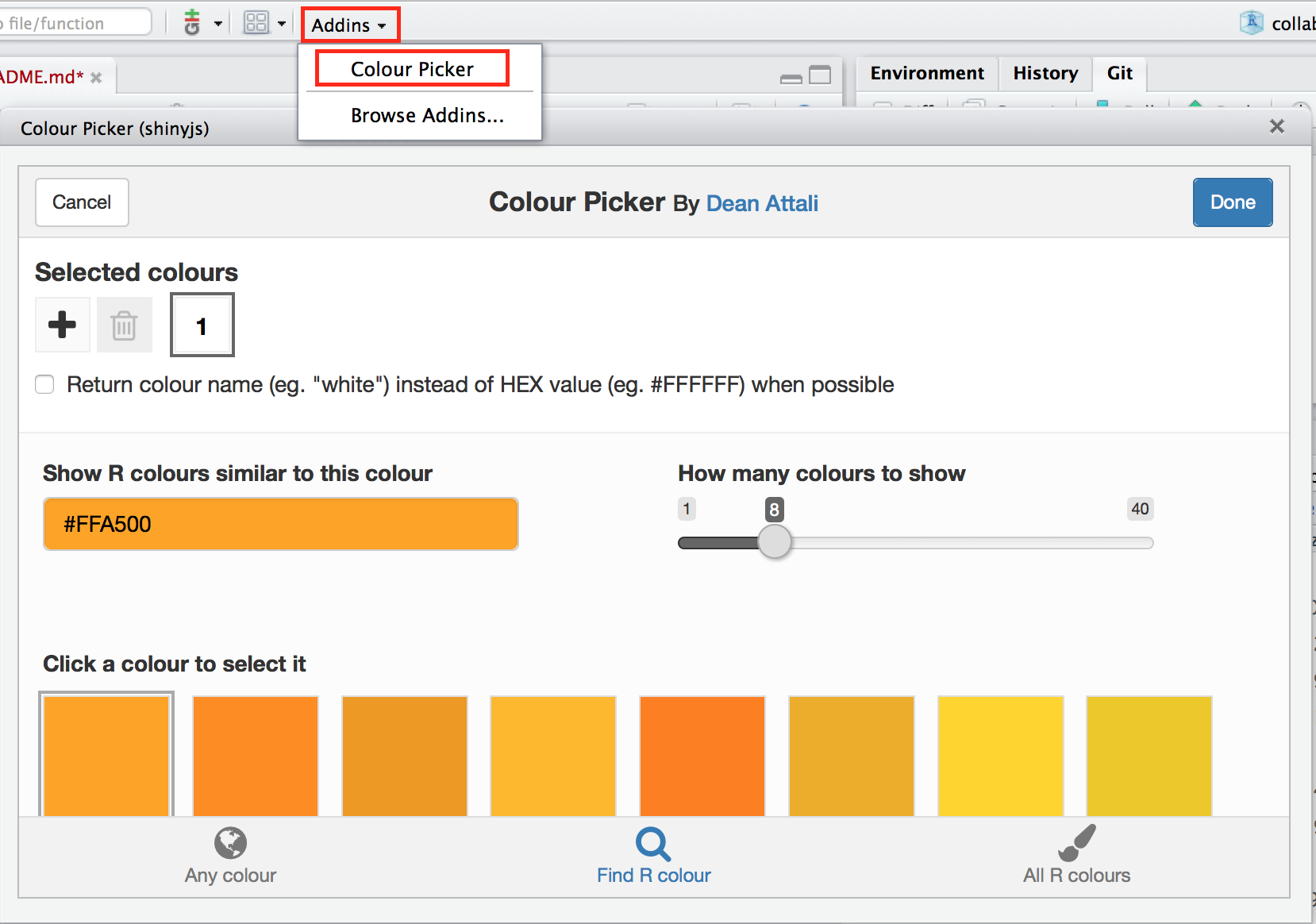
color: e.g."red","#FFFFFF"(tip: pick a color using Rstudio's Color picker:)
-
Use the git tab, tick the box next to your new script ONLY and commit.
-
Supply a descriptive message of the commit.
- push your changes to github
- create a pull request to merge your changes to the master repo
- most likely it'll be a thumbs up! But I might also request a correction if I spot an error.
Once all parameters are collated, look out for the results!
You will need R and Rstudio installed on your local system.
First, open Rstudio and install a couple of setup packages you will need.
install.packages(c("renv", "usethis"))
Next, clone your own fork of the repository. In the code example below you will need to edit the repo_spec argument with your own GitHub account name as well as supply the path where you want the repo cloned to in destdir.
usethis::create_from_github(repo_spec = "YOUR-ACCOUNT-NAME/collaborative_github_exercise",
fork = FALSE, destdir = "path/where/you/want/the/repo/cloned/to")This will clone your fork from GitHub and open up the project.
The project uses renv for dependency management. To install all required dependencies into the local project library run:
renv::restore()Next, add the RSE-Sheffield/collaborative_github_exercise remote repo to list of remotes as upstream
usethis::use_git_remote(name = "upstream", url ="https://github.com/RSE-Sheffield/collaborative_github_exercise.git" , overwrite = TRUE)Next, pull from the upstream repository:
usethis::pr_merge_main()You should now have all the parameter files submitted by participants in your local params/ folder
Finally, to generate the Evolottery html webpage, you will need to knit the plot_trait_evolution.Rmd file.
Open the plot_trait_evolution.Rmd file and on the top panel click on the knit button. This runs the code and renders the content of the file to html.
Note that because trait evolution is random, your version of the results will differ from those published in the upstream repo.