This is a Pytorch implementation of
SS-GNN, a simple-structured GNN model for drug-target binding affinity (DTBA) prediction as described in the following paper:
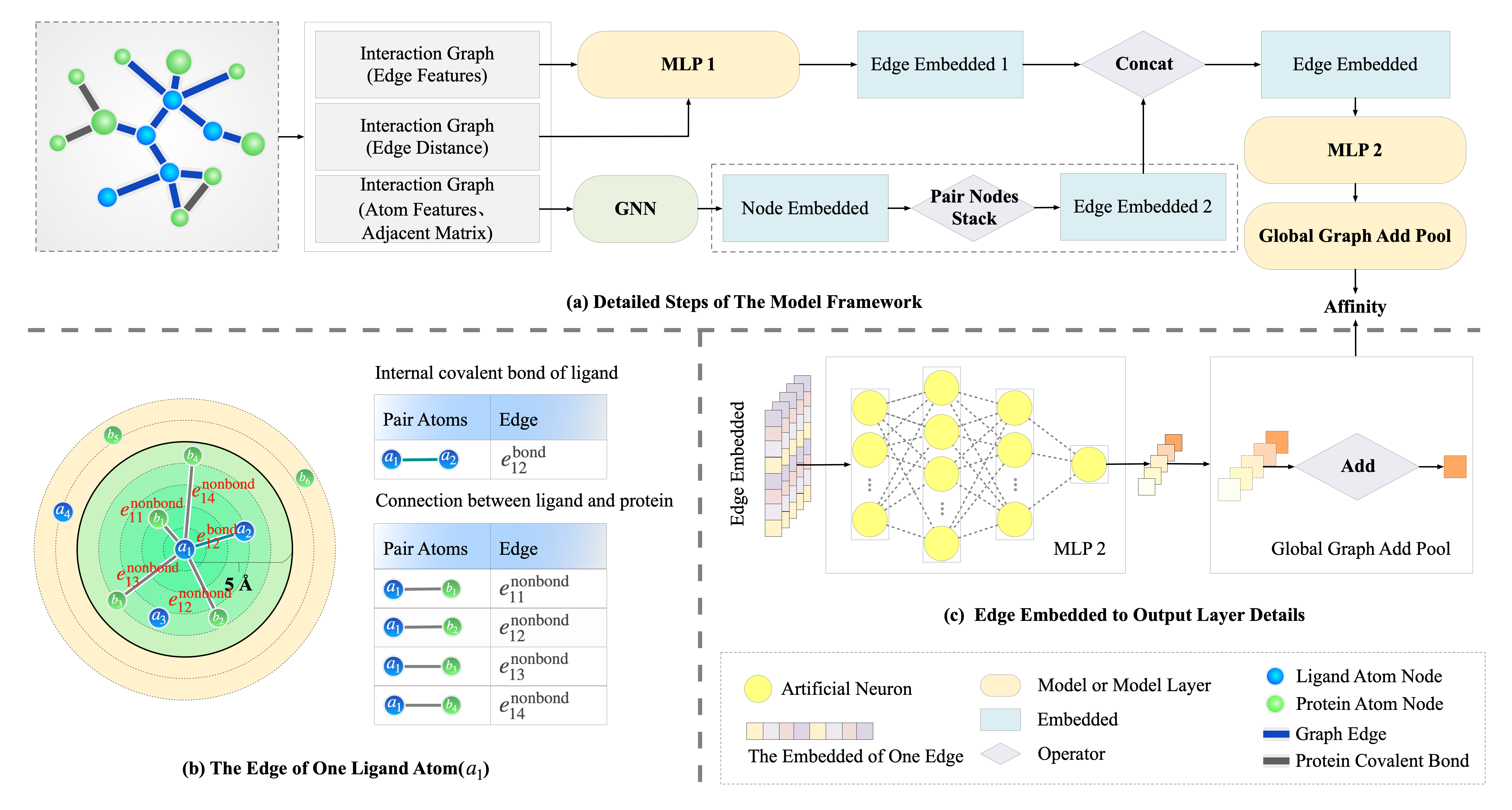
The SS-GNN defines the prediction of DTBA as a regression task, in which the model’s input is the drug-target representation, and the output is a continuous value representing the binding affinity score between the drug and the target protein. The overall architecture of the SS-GNN is shown in the figure below.
-
Setup
Necessary packages should be installed to run the SS-GNN model. Dependecies:
- python >= 3.7
- Pytorch (>=1.6.0),
- numpy,
- scipy,
- scikit-learn.
-
Datasets
We adopt the PDBbind dataset v2019 for experiments and employ two test sets (the v2016 and v2013 core sets) to test the performance of SS-GNN.
-
Train the model
Use the
train.pyscript to train the model.
Please cite the following paper if you find this repository useful.