'''
OSGA-V for optimal data fusion in presence of unknown correlations
'''
import numpy as np
import math
from IPython.core.debugger import set_trace
def to_matrix_vector(matrix_array):
'''
Converts an array of A_i to [A_1, ..., A_N]^T
'''
n, m, l = matrix_array.shape
assert (m == l)
return np.transpose(np.concatenate(matrix_array, axis=1))
def to_matrix_array(matrix_vector):
'''
Converts a matrix vector to an array of matrices
'''
n, m = matrix_vector.shape
assert (n > m)
return np.asarray(np.hsplit(np.transpose(matrix_vector), n / m))
def pus(delta, alpha_max, kappa_p, kappa, alpha, h_bar, gamma_bar, eta_bar, w_bar, eta, h, gamma, w):
'''
Performs the parameter updating scheme
'''
R = (eta - eta_bar) / (delta * alpha * eta)
if R < 1:
alpha_bar = alpha * math.exp(-kappa)
else:
alpha_bar = min(alpha * math.exp(kappa_p * (R - 1)), alpha_max)
alpha = alpha_bar
if eta_bar < eta:
h = h_bar
gamma = gamma_bar
eta = eta_bar
w = w_bar
return alpha, h, gamma, eta, w
def dagger_transform(in_mat_vec):
'''
Converts a matrix vector to its daggger edition (element-matrix-wise transpose)
'''
n, m = in_mat_vec.shape
splitted = np.asarray(np.hsplit(np.transpose(in_mat_vec), n/m))
for i in range(len(splitted)):
splitted[i] = np.transpose(splitted[i])
return to_matrix_vector(splitted)
def solve_subproblem(gamma_b, h, b_hat, Sigma_inv, V_prime_joint_neg_sqrt):
'''
Solves the E(gamma_b, h), U(gamma_b, h) subproblem
All vectors are assumed to be in matrix vector format
'''
NM, M = h.shape # shape NM x M
assert (NM > M and (NM % M) == 0)
N = int(NM / M)
assert ((NM, M) == b_hat.shape)
assert ((M, M) == Sigma_inv.shape)
assert ((NM, NM) == V_prime_joint_neg_sqrt.shape)
h_hat = np.matmul(V_prime_joint_neg_sqrt, h)
Omega = np.matmul(np.transpose(h_hat), b_hat)
Omega_Sigma_inv = np.matmul(Omega, Sigma_inv)
b_hat_array = to_matrix_array(b_hat)
h_hat_array = to_matrix_array(h_hat)
r_list = list()
for i in range(N):
r_list.append(np.matmul(Omega_Sigma_inv, b_hat_array[i]) - h_hat_array[i])
r_array = np.asarray(r_list)
beta1 = 1 + np.trace(Sigma_inv) / 2
beta2 = gamma_b
for i in range(N):
b_i_T_sigma_inv = np.matmul(np.transpose(b_hat_array[i]), Sigma_inv)
beta2 = beta2 + np.trace(np.matmul(r_array[i], b_i_T_sigma_inv))
beta2 = beta2 + np.trace(np.matmul(h_hat_array[i], b_i_T_sigma_inv))
beta3 = 0
for i in range(N):
beta3 = beta3 + np.trace(np.matmul(r_array[i], np.transpose(r_array[i]))) / 2
beta3 = beta3 + np.trace(np.matmul(h_hat_array[i], np.transpose(r_array[i])))
eta = (-beta2 + np.sqrt(beta2 * beta2 - 4 * beta1 * beta3)) / (2 * beta1)
p_list = list()
for i in range(N):
p_list.append(r_array[i] / eta + np.matmul(Sigma_inv, b_hat_array[i]))
p_mat_vec = to_matrix_vector(np.asarray(p_list))
u_hat_mat_vec = np.matmul(V_prime_joint_neg_sqrt, p_mat_vec)
return eta, u_hat_mat_vec
def oracle_call(x, unknown_corr_flags, V_prime):
'''
Given the value point x, given as a matrix vector,
return a subgradient, and f(x)
V_prime has shape (N, N, M, M) having I for diagonal blocks and
V_prime_ij for known correlations
'''
xa = to_matrix_array(x)
N, M, _ = xa.shape
fx = 0
sub_gradient = np.zeros((N, M, M), np.float32)
for i in range(N):
for j in range(N):
if j != i and unknown_corr_flags[i, j]:
C, Lambda_ij, D = np.linalg.svd(np.matmul(np.transpose(xa[j]), xa[i]))
D = np.transpose(D) # convention of python linalg library
fx = fx + np.sum(Lambda_ij)
sub_gradient[i] = sub_gradient[i] + np.matmul(np.matmul(xa[j], C), np.transpose(D))
else:
sub_gradient[i] = sub_gradient[i] + np.matmul(xa[j], np.transpose(V_prime[i, j]))
fx = fx + np.trace(np.matmul(xa[i], np.matmul(V_prime[i, j], np.transpose(xa[j]))))
sub_gradient[i] = sub_gradient[i] * 2.0
return sub_gradient, fx
# Usage:
# a_i, mse, v_opt, v_ij_opt = gdof (
# N, M,
# joint_covariance,
# unknown_index_matrix)
#
# Inputs:
# N - the input measurement count
# M - the input measurement dimension
# joint_covariance - the joint covariance matrix
# for the all the measurements. It's a 4-d
# tensor, with the first two dimensions
# referring to the measurements and the last
# two dimensions referring to the measurement
# components. For unknown cross-correlation
# matrices, the values are not used.
# unknown_corr_flags - a bool numpy array.
# the element of the matrix at
# location (i,j) is set to be one if V_ij is unknown.
# otherwise it is set to be zero.
#
# Outputs:
# . a_i - the matrix weights, a tensor of 3d, with the first
# . dimension index being the measurement index
# . mse - the resulting mean square error
# . v_opt - the estimate covariance
# v_ij_opt - the maximizing cross correlation matrix at (i,j)
def OSGAV(N, M, joint_covariance, unknown_corr_flags, max_iteration=1000):
assert (N, N, M, M) == joint_covariance.shape
assert (N, N) == unknown_index_matrix.shape
mu = 0
alpha_max = 0.5
delta = 0.5
kappa_p = 0.4
kappa = 0.8
V = joint_covariance
B = np.zeros((N, M, M), dtype=np.float32)
Lambda_inv_sqrt = np.zeros((N,M), dtype=np.float32)
U = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
Lambda_inv_sqrt[i], U[i] = np.linalg.eigh(joint_covariance[i, i])
Lambda_inv_sqrt[i] = np.reciprocal(np.sqrt(Lambda_inv_sqrt[i]))
B[i] = np.matmul(np.diag(Lambda_inv_sqrt[i]), np.transpose(U[i]))
V_prime = np.zeros((N, N, M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i == j:
V_prime[i, j] = np.identity(M, dtype=np.float32)
elif (not unknown_corr_flags[i, j]):
V_prime[i, j] = np.matmul(np.diag(Lambda_inv_sqrt[i]), np.transpose(U[i]))
V_prime[i, j] = np.matmul(V_prime[i, j], V[i, j])
V_prime[i, j] = np.matmul(V_prime[i, j], U[j])
V_prime[i, j] = np.matmul(V_prime[i, j], np.diag(Lambda_inv_sqrt[j]))
V_joint_list = list()
for i in range(N):
V_joint_list.append(np.concatenate(V_prime[i], axis=1))
V_prime_joint = np.concatenate(np.asarray(V_joint_list), axis=0)
lambda_v, Ut = np.linalg.eigh(V_prime_joint)
V_prime_joint_neg_sqrt = np.matmul(np.matmul(Ut, np.diag(1.0 / np.sqrt(lambda_v))), np.transpose(Ut))
B_mat_vec = to_matrix_vector(B)
B_hat_mat_vec = np.matmul(V_prime_joint_neg_sqrt, dagger_transform(B_mat_vec))
Sigma = np.matmul(np.transpose(B_hat_mat_vec), B_hat_mat_vec)
Sigma_inv = np.linalg.inv(Sigma)
x_opt = np.array([[ 0., 1.],[0, 0], [ 1.41421354, 0.], [0, 0],[0, 0],[0, 0]], dtype=np.float32)
# initial A_prime values
A_prime = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
A_prime[i] = np.matmul(U[i], np.diag(np.reciprocal(Lambda_inv_sqrt[i]))) / N
x_b = to_matrix_vector(A_prime)
sub_gradient, fx_b = oracle_call(x_b, unknown_corr_flags, V_prime)
h = to_matrix_vector(sub_gradient)
gamma = fx_b - np.trace(np.matmul(np.transpose(h), x_b)) # - <h, xb>
gamma_b = gamma - fx_b
eta, w = solve_subproblem(gamma_b, h, B_hat_mat_vec, Sigma_inv, V_prime_joint_neg_sqrt)
eta = eta - mu
alpha = alpha_max
mse_iteration = []
for iteration in range(max_iteration):
x = x_b + alpha * (w - x_b)
sub_gradient, fx = oracle_call(x, unknown_corr_flags, V_prime)
g = to_matrix_vector(sub_gradient)
h_bar = h + alpha * (g - h)
gamma_bar = gamma + alpha * (fx - np.trace(np.matmul(np.transpose(g), x)) - gamma)
if fx < fx_b:
x_b_prime = x
fx_b_prime = fx
else:
x_b_prime = x_b
fx_b_prime = fx_b
gamma_b_prime = gamma_bar - fx_b_prime
eta_prime, w_prime = solve_subproblem(gamma_b_prime, h_bar, B_hat_mat_vec, Sigma_inv, V_prime_joint_neg_sqrt)
if eta_prime < mu:
set_trace()
eta_prime, w_prime = solve_subproblem(gamma_b_prime, h_bar, B_hat_mat_vec, Sigma_inv, V_prime_joint_neg_sqrt)
eta_prime = eta_prime - mu
x_prime = x_b_prime + alpha * (w_prime - x_b_prime)
_, fx_prime = oracle_call(x_prime, unknown_corr_flags, V_prime)
if fx_b_prime < fx_prime:
x_b = x_b_prime
fx_b = fx_b_prime
else:
x_b = x_prime
fx_b = fx_prime
alpha, h, gamma, eta, w = pus(
delta, alpha_max, kappa_p, kappa, alpha, h_bar, gamma_bar, eta_prime, w_prime, eta, h, gamma, w)
#print('Iteration', iteration, 'fx_b', fx_b, 'eta', eta)
mse_iteration.append(fx_b)
A_prime_best = to_matrix_array(x_b)
A = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
A[i] = np.matmul(np.matmul(A_prime_best[i], np.diag(Lambda_inv_sqrt[i])), np.transpose(U[i]))
V_ij_opt = np.zeros((N, N, M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and unknown_index_matrix[i, j]:
C_ij, Lambda_ij, D_ij = np.linalg.svd(np.matmul(np.transpose(A_prime_best[j]), A_prime_best[i]))
part_left = np.matmul(U[i], np.diag(np.reciprocal(Lambda_inv_sqrt[i])))
part_middle = np.matmul(np.transpose(D_ij), np.transpose(C_ij))
part_right = np.matmul(np.diag(np.reciprocal(Lambda_inv_sqrt[j])), np.transpose(U[j]))
V_ij_opt[i, j] = np.matmul(np.matmul(part_left, part_middle), part_right)
V_opt = np.zeros((M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and unknown_index_matrix[i, j]:
V_opt = V_opt + np.matmul(np.matmul(A[i], V_ij_opt[i, j]), np.transpose(A[j]))
else:
V_opt = V_opt + np.matmul(np.matmul(A[i], V[i, j]), np.transpose(A[j]))
return A, fx_b, V_opt, V_ij_opt, mse_iterationfrom io import StringIO
from unittest import *
class TestOSGAV(TestCase):
def test_matrix_conversion(self):
a = [[1,2],[3,4]]
b = [[5,6],[7,8]]
c = [[9, 10], [11, 12]]
s = np.array(np.stack((a, b, c), axis=0), np.float32)
v = to_matrix_vector(s)
sp = to_matrix_array(v)
self.assertTrue((s == sp).all())
self.assertTrue((v == np.array([[1, 3], [2, 4], [5, 7], [6, 8], [9, 11], [10, 12]], np.float32)).all())
def test_pus(self):
alpha_max = 0.5
delta = 0.5
kappa_p = 0.4
kappa = 0.8
alpha = alpha = 0.1
eta = 0.3
h_bar = [1, 2]
gamma_bar = 1
gamma = 2
eta_bar = 0.4
w_bar = [3, 4]
w = [5, 6]
h = [7, 8]
w = [9, 10]
delta = 0.5
alpha_new, h_new, gamma_new, eta_new, w_new = pus(
delta, alpha_max, kappa_p, kappa, alpha, h_bar, gamma_bar, eta_bar, w_bar, eta, h, gamma, w)
self.assertTrue(alpha > alpha_new)
self.assertEqual(eta_new, eta)
self.assertEqual(h_new, h)
self.assertEqual(gamma_new, gamma)
self.assertEqual(w_new, w)
eta_bar = 0.2
alpha_new, h_new, gamma_new, eta_new, w_new = pus(
delta, alpha_max, kappa_p, kappa, alpha, h_bar, gamma_bar, eta_bar, w_bar, eta, h, gamma, w)
self.assertTrue(alpha < alpha_new)
self.assertEqual(eta_new, eta_bar)
self.assertEqual(h_new, h_bar)
self.assertEqual(gamma_new, gamma_bar)
self.assertEqual(w_new, w_bar)
def test_dagger_transform(self):
m = np.array([[1, 2], [3, 4], [5, 6], [7, 8], [9, 10], [11, 12]], np.float32)
n = np.array([[[1, 3], [2, 4], [5, 7], [6, 8], [9, 11], [10, 12]]], np.float32)
self.assertTrue((n == dagger_transform(m)).all())
def test_solve_subproblem(self):
'''
Here we use a special problem to cross check with solve_subproblem:
maiximize {-(r + <h, x>) / ((1/2) x^T V x + 1)} subject to a^T x = 1
using Lagrange multiplier, with the solution u and e,
Let y = <h, u>, z = <Vu, u>, lambda = - y / (1 + z / 2) + z (r + y) / (1 + z/2)^2
then we must have
- h/(1 + z / 2) + (r + y)/(1 + z / 2)^2 * Vu - lambda a = 0
'''
N = 3
M = 1
V_prime_joint = np.array([[3, 2, 1], [2, 5, 0], [1, 0, 1]], dtype=np.float32)
lambda_v, U = np.linalg.eigh(V_prime_joint)
V_prime_joint_neg_sqrt = np.matmul(np.matmul(U, np.diag(1.0 / np.sqrt(lambda_v))), np.transpose(U))
B_mat_vec = np.reshape(1 / np.sqrt(lambda_v), (N * M, M))
B_hat_mat_vec = np.matmul(V_prime_joint_neg_sqrt, B_mat_vec)
Sigma = np.matmul(np.transpose(B_hat_mat_vec), B_hat_mat_vec)
Sigma_inv = np.linalg.inv(Sigma)
gamma_b = 1.0
h = np.array([[1], [2], [3]], dtype=np.float32)
eta, u_hat_mat_vec = solve_subproblem(gamma_b, h, B_hat_mat_vec, Sigma_inv, V_prime_joint_neg_sqrt)
y = np.matmul(np.transpose(u_hat_mat_vec), h)[0, 0]
vu = np.matmul(V_prime_joint, u_hat_mat_vec)
z = np.matmul(np.transpose(vu), u_hat_mat_vec)[0, 0]
eta2 = -(gamma_b + y) / (1 + z / 2)
self.assertTrue(abs(eta - eta2) < 1.0e-5)
constraint = np.matmul(np.transpose(u_hat_mat_vec), B_mat_vec)[0, 0]
self.assertTrue(abs(constraint - 1.0) < 1.0e-5)
lambda_v = -y / (1 + z / 2) + z * (gamma_b + y) / (1 + z / 2) / (1 + z / 2)
a_vec = np.reshape(B_mat_vec, (N))
h_vec = np.reshape(h, (N))
vu = np.reshape(vu, (N))
residue = abs(- h_vec / (1 + z / 2) + (gamma_b + y) / (1 + z / 2) / (1 + z / 2) * vu - lambda_v * a_vec)
self.assertTrue((residue < 1.0e-5).all())
t = TestOSGAV()
suite = TestLoader().loadTestsFromModule(t)
TextTestRunner().run(suite)....
----------------------------------------------------------------------
Ran 4 tests in 0.017s
OK
<unittest.runner.TextTestResult run=4 errors=0 failures=0>
# Test case 1 from Spyridon Leonardos and Kostas Daniilidis
import time
x1 = np.array([0.0, 0.0], np.float32)
V11 = np.array([[5.0, 0], [0, 1.0]], np.float32)
x2 = np.array([0.0, 0.0], np.float32)
V22 = np.array([[2.0, 0], [0, 7.0]], np.float32)
x3 = np.array([0.0, 0.0], np.float32)
V33 = np.array([[4.0, 0], [0, 100.0]], np.float32)
joint_covariance = np.array([[V11, V11, V11],[V22, V22, V22], [V33, V33, V33]], np.float32)
unknown_index_matrix = np.array([[False, True, True], [True, False, True], [True, True, False]], dtype=bool)
N = 3
M = 2
iteration_count = 110
start_time = time.time()
a_i, mse, v_opt, v_ij_opt, osgav_css_iteration = OSGAV(N, M, joint_covariance, unknown_index_matrix, max_iteration=iteration_count)
osgav_elapsed_time = time.time() - start_time
print('\nOSGA-V -', iteration_count, 'iteration elapsed time: ', osgav_elapsed_time)
print('OSGA-V - MSE: ', mse, '\n')
start_time = time.time()
a_i, mse, v_opt, v_ij_opt, osgav_css_iteration = OSGAV(N, M, joint_covariance, unknown_index_matrix)
osgav_elapsed_time = time.time() - start_time
print('OSGA-V - Elapsed time: ', osgav_elapsed_time)
print('OSGA-V - A_i: ', a_i)
print('OSGA-V - MSE: ', mse)
print('OSGA-V - Covariance: ', v_opt)OSGA-V - 110 iteration elapsed time: 0.14423632621765137
OSGA-V - MSE: 3.0034182314
OSGA-V - Elapsed time: 1.5090413093566895
OSGA-V - A_i: [[[ 2.05970014e-06 -5.54971358e-08]
[ 9.15520104e-08 9.99999344e-01]]
[[ 9.99997377e-01 -1.57472584e-07]
[ -1.02756132e-07 1.68238230e-07]]
[[ 7.99670943e-07 2.12969312e-07]
[ 1.12004805e-08 2.42156322e-08]]]
OSGA-V - MSE: 3.00001014186
OSGA-V - Covariance: [[ 2.00001025 -0.04545899]
[-0.04545899 1.00000012]]
For comparison purpose. Codes are adapted from PSOF paper codes.
import numpy as np
import math
def func_constant_step_size(sub_gradient, iteration, diff_from_best_estimate):
return 0.0004
def func_constant_step_length(sub_gradient, iteration, diff_from_best_estimate):
global csl_initial_norm
N,M,_ = sub_gradient.shape
norm = 0
for i in range(N):
norm += np.trace(np.matmul(sub_gradient[i], np.transpose(sub_gradient[i])))
return 0.002 / np.sqrt(norm)
def func_square_summable_not_summable(sub_gradient, iteration, diff_from_best_estimate):
return 3.0 / (iteration + 1)
def func_not_summable_diminishing_step_size(sub_gradient, iteration, diff_from_best_estimate):
return 0.035 / np.sqrt(iteration + 1)
def func_not_summable_diminishing_step_length(sub_gradient, iteration, diff_from_best_estimate):
global nsdsl_initial_norm
N,M,_ = sub_gradient.shape
norm = 0
for i in range(N):
norm += np.trace(np.matmul(sub_gradient[i], np.transpose(sub_gradient[i])))
return 0.19 / np.sqrt(norm * (iteration + 1))
def func_polyak_with_estimate(sub_gradient, iteration, diff_from_best_estimate):
global polyak_initial_norm
N,M,_ = sub_gradient.shape
norm = 0
for i in range(N):
norm += np.trace(np.matmul(sub_gradient[i], np.transpose(sub_gradient[i])))
return (0.035 * norm / np.sqrt(iteration + 1) + diff_from_best_estimate) / norm
# Projected Subgradient Method
# Usage:
# a_i, mse, v_opt, v_ij_opt = gdof (
# N, M,
# joint_covariance,
# unknown_index_matrix)
#
# Inputs:
# N - the input measurement count
# M - the input measurement dimension
# joint_covariance - the joint covariance matrix
# for the all the measurements. It's a 4-d
# tensor, with the first two dimensions
# referring to the measurements and the last
# two dimensions referring to the measurement
# components. For unknown cross-correlation
# matrices, the values are not used.
# unknown_index_matrix - a bool numpy array.
# the element of the matrix at
# location (i,j) is set to be one if V_ij is unknown.
# otherwise it is set to be zero.
#
# Outputs:
# . a_i - the matrix weights, a tensor of 3d, with the first
# . dimension index being the measurement index
# . mse - the resulting mean square error
# . v_opt - the estimate covariance
# v_ij_opt - the maximizing cross correlation matrix at (i,j)
def PSOF(N, M, joint_covariance, unknown_index_matrix, step_func=func_constant_step_size, max_iteration=12000):
assert (N, N, M, M) == joint_covariance.shape
assert (N, N) == unknown_index_matrix.shape
V = joint_covariance
B = np.zeros((N, M, M), dtype=np.float32)
Lambda_inv_sqrt = np.zeros((N,M), dtype=np.float32)
U = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
Lambda_inv_sqrt[i], U[i] = np.linalg.eigh(joint_covariance[i, i])
Lambda_inv_sqrt[i] = np.reciprocal(np.sqrt(Lambda_inv_sqrt[i]))
B[i] = np.matmul(np.diag(Lambda_inv_sqrt[i]), np.transpose(U[i]))
V_prime = np.zeros((N, N, M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and (not unknown_index_matrix[i, j]):
V_prime[i, j] = np.matmul(np.diag(Lambda_inv_sqrt[i]), np.transpose(U[i]))
V_prime[i, j] = np.matmul(V_prime[i, j], V[i, j])
V_prime[i, j] = np.matmul(V_prime[i, j], U[j])
V_prime[i, j] = np.matmul(V_prime[i, j], np.diag(Lambda_inv_sqrt[j]))
Sigma = np.zeros((M, M), dtype=np.float32)
for i in range(N):
Sigma = Sigma + np.linalg.inv(V[i, i])
Sigma_inv = np.linalg.inv(Sigma)
# initial A_prime values
A_prime = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
A_prime[i] = np.matmul(U[i], np.diag(np.reciprocal(Lambda_inv_sqrt[i]))) / N
epislon = 1.0e-12
mse = np.finfo(np.float32).max
mse_iteration = []
mse_best = np.finfo(np.float32).max
last_mse_diff = 0
A_prime_best = np.zeros((N, M, M), dtype=np.float32)
for iteration in range(max_iteration):
# Get SVD of A[j]^T A[i]
C = np.zeros((N, N, M, M), dtype=np.float32)
D = np.zeros((N, N, M, M), dtype=np.float32)
Lambda_ij = np.zeros((N, N, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and unknown_index_matrix[i, j]:
C[i, j], Lambda_ij[i, j], D[i, j] = np.linalg.svd(np.matmul(np.transpose(A_prime[j]), A_prime[i]))
D[i, j] = np.transpose(D[i, j]) # convention of python linalg library
# compute subgradients
dA_prime = np.zeros((N, M, M), np.float32)
for i in range(N):
dA_prime[i] = A_prime[i]
for j in range(N):
if j != i:
if unknown_index_matrix[i, j]:
dA_prime[i] = dA_prime[i] + np.matmul(np.matmul(A_prime[j], C[i, j]), np.transpose(D[i, j]))
else:
dA_prime[i] = dA_prime[i] + np.matmul(A_prime[j], np.transpose(V_prime[i, j]))
dA_prime[i] = dA_prime[i] * 2.0
# apply step size & subgradient
step = step_func(dA_prime, iteration, last_mse_diff)
for i in range(N):
A_prime[i] = A_prime[i] - dA_prime[i] * step
# project onto the constraint hyperplanes
A_prime_dot_B = np.zeros((M, M), np.float32)
for i in range(N):
A_prime_dot_B = A_prime_dot_B + np.matmul(A_prime[i], B[i])
A_prime_dot_B_Sigma_inv = np.matmul(np.eye(M, dtype=np.float32) - A_prime_dot_B, Sigma_inv)
for i in range(N):
A_prime[i] = A_prime[i] + np.matmul(A_prime_dot_B_Sigma_inv, np.transpose(B[i]))
# compute mse
mse_prime = 0.0
for i in range(N):
mse_prime = mse_prime + np.trace(np.matmul(np.transpose(A_prime[i]), A_prime[i]))
for i in range(N):
for j in range(N):
if i != j:
if unknown_index_matrix[i, j]:
_, sigmas, _ = np.linalg.svd(np.matmul(np.transpose(A_prime[j]), A_prime[i]))
mse_prime = mse_prime + np.sum(sigmas)
else:
mse_prime = mse_prime + np.trace(np.matmul(np.matmul(A_prime[i], V_prime[i, j]), np.transpose(A_prime[j])))
mse_iteration.append(mse_prime)
#print('mse: ', mse_prime, mse_best, sigmas, A_prime)
if mse_prime < mse_best:
mse_best = mse_prime
A_prime_best = A_prime
last_mse_diff = 0
last_mse_diff = mse_prime - mse_best
A = np.zeros((N, M, M), dtype=np.float32)
for i in range(N):
A[i] = np.matmul(np.matmul(A_prime_best[i], np.diag(Lambda_inv_sqrt[i])), np.transpose(U[i]))
V_ij_opt = np.zeros((N, N, M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and unknown_index_matrix[i, j]:
C_ij, Lambda_ij, D_ij = np.linalg.svd(np.matmul(np.transpose(A_prime_best[j]), A_prime_best[i]))
part_left = np.matmul(U[i], np.diag(np.reciprocal(Lambda_inv_sqrt[i])))
part_middle = np.matmul(np.transpose(D_ij), np.transpose(C_ij))
part_right = np.matmul(np.diag(np.reciprocal(Lambda_inv_sqrt[j])), np.transpose(U[j]))
V_ij_opt[i, j] = np.matmul(np.matmul(part_left, part_middle), part_right)
V_opt = np.zeros((M, M), dtype=np.float32)
for i in range(N):
for j in range(N):
if i != j and unknown_index_matrix[i, j]:
V_opt = V_opt + np.matmul(np.matmul(A[i], V_ij_opt[i, j]), np.transpose(A[j]))
else:
V_opt = V_opt + np.matmul(np.matmul(A[i], V[i, j]), np.transpose(A[j]))
return A, mse_best, V_opt, V_ij_opt, mse_iteration# Test case 1 from Spyridon Leonardos and Kostas Daniilidis
import time
x1 = np.array([0.0, 0.0], np.float32)
V11 = np.array([[5.0, 0], [0, 1.0]], np.float32)
x2 = np.array([0.0, 0.0], np.float32)
V22 = np.array([[2.0, 0], [0, 7.0]], np.float32)
x3 = np.array([0.0, 0.0], np.float32)
V33 = np.array([[4.0, 0], [0, 100.0]], np.float32)
joint_covariance = np.array([[V11, V11, V11],[V22, V22, V22], [V33, V33, V33]], np.float32)
unknown_index_matrix = np.array([[False, True, True], [True, False, True], [True, True, False]], dtype=bool)
N = 3
M = 2
iteration_count = 500
start_time = time.time()
a_i, mse, v_opt, v_ij_opt, mse_nsdss_iteration = PSOF(N, M, joint_covariance, unknown_index_matrix, step_func=func_not_summable_diminishing_step_size, max_iteration=iteration_count)
nsdss_elapsed_time = time.time() - start_time
print('\nNSDSS -', iteration_count, 'iteration elapsed time: ', nsdss_elapsed_time)
print('NSDSS - MSE: ', mse, '\n')
start_time = time.time()
a_i, mse, v_opt, v_ij_opt, mse_nsdss_iteration = PSOF(N, M, joint_covariance, unknown_index_matrix, step_func=func_not_summable_diminishing_step_size)
nsdss_elapsed_time = time.time() - start_time
print('NSDSS - Elapsed time: ', nsdss_elapsed_time)
print('NSDSS - A_i: ', a_i)
print('NSDSS - MSE: ', mse)
print('NSDSS - Covariance: ', v_opt)NSDSS - 500 iteration elapsed time: 0.33220958709716797
NSDSS - MSE: 3.00437980058
NSDSS - Elapsed time: 7.936528444290161
NSDSS - A_i: [[[ 7.98037145e-05 -4.49721119e-06]
[ 1.72876062e-05 9.99835968e-01]]
[[ 9.99588013e-01 4.02844807e-06]
[ 2.73404785e-05 1.13685725e-04]]
[[ 3.32220545e-04 4.68767041e-07]
[ -4.46280828e-05 5.03705596e-05]]]
NSDSS - MSE: 3.00057295242
NSDSS - Covariance: [[ 2.00073457 0.08226774]
[ 0.08226774 1.00130045]]
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
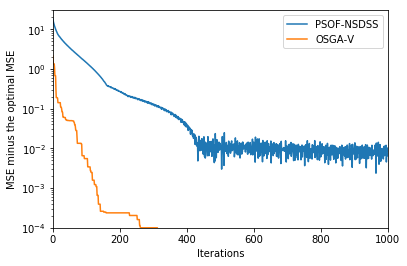
plt.ylim((1.0e-4, 30.0))
plt.xlim((0.0, 1000.0))
plt.yscale('log')
plt.plot(np.array(mse_nsdss_iteration, dtype=np.float32) - 3.0, label="PSOF-NSDSS")
plt.plot(np.array(osgav_css_iteration, dtype=np.float32) - 3.0, label="OSGA-V")
plt.ylabel('MSE minus the optimal MSE')
plt.xlabel('Iterations')
plt.legend()
plt.figure()
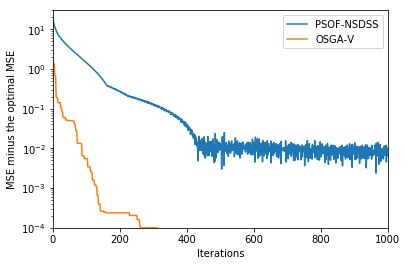
plt.ylim((1.0e-4, 30.0))
plt.xlim((0.0, 1000.0))
plt.yscale('log')
plt.plot(np.array(mse_nsdss_iteration, dtype=np.float32) - 3.0, label="PSOF-NSDSS")
plt.plot(np.array(osgav_css_iteration, dtype=np.float32) - 3.0, label="OSGA-V")
plt.ylabel('MSE minus the optimal MSE')
plt.xlabel('Iterations')
plt.legend()<matplotlib.legend.Legend at 0x7f15b8d60ba8>