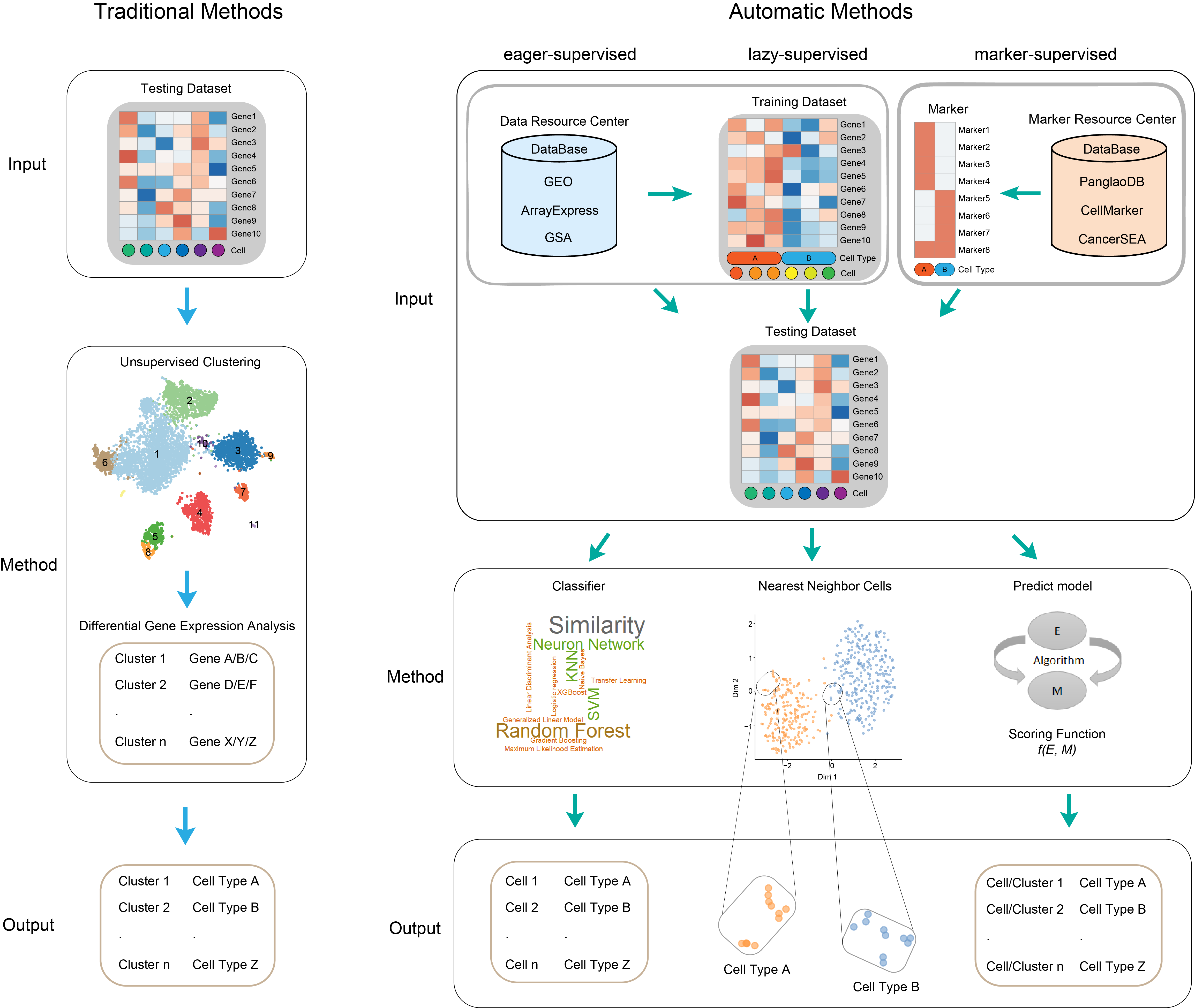
AutomaticCellTypeIdentification is a wrapper of published automatic cell type identification methods which contains supervised methods, unsupervised methods and semi-supervised methods.
You can install AutomaticCellTypeIdentification from github with:
devtools::install_github('xiebb123456/AutomaticCellTypeIdentification')Note: AutomaticCellTypeIdentification is a wrapper of published methods, the needed package is in Description file.
sudo docker pull registry.cn-hangzhou.aliyuncs.com/xiebb123456/automaticcelltypeidentification
Now, three interface of eagersupervised, lazysupervised, markersupervised methods supports the available automatic methods.
eagersupervised methods include ACTINN, CaSTLe, CHETAH, clustifyr, Garnett, Markercount, MARS, scClassifR, scHPL, SciBet, scID, scLearn, scmapcluster, scPred, scVI, Seurat, SingleCellNet and SingleR.
lazysupervised methods include CELLBLAST and scmapcell.
markersupervised methods include scTyper, Markercount, SCSA, DigitalCellSorter and SCINA.
The input of training and testing data is count matrix, the row is gene and the column is cell.
eagersupervised(train,test,label_train,method='Seurat')lazysupervised(train,test,label_train,method='CELLBLAST')markersupervised(test,marker,method='SCSA')Note: the conda environment of python-based method is needed to load at the beginning in R.
For more details and basic usage see following tutorials: Guided Tutorial
Feel free to submit an issue or contact us at xiebb7@mail.sysu.edu.cn for problems about the package installation and usage.