Multi-modal Physiological Signals based Squeeze-and-Excitation Network with Domain Adversarial Learning for Sleep Staging
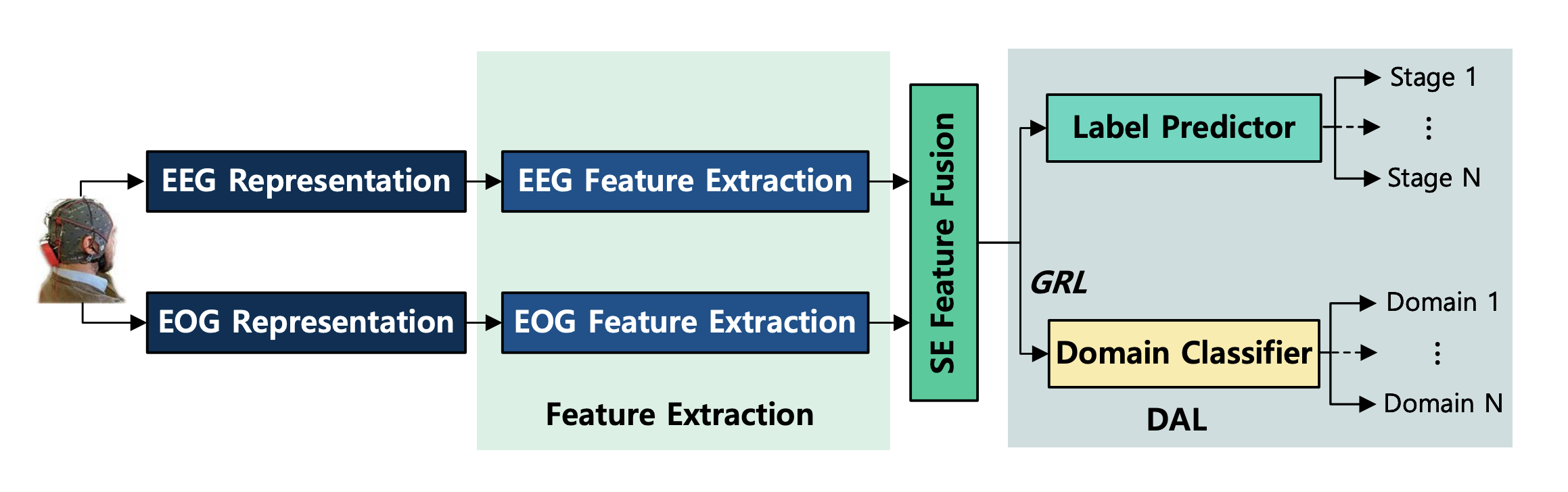
The multi-modal physiological signals based Squeeze-and-Excitation Network with Domain Adversarial Learning (SEN-DAL) is a deep learning model for sleep staging. The SEN-DAL is made up of two independent feature extraction networks for modeling the heterogeneity, a Multi-modal Squeeze-and-Excitation feature fusion module for adaptively utilizing the multi-modal signals, and a Domain Adversarial Learning module to extract subject-invariant sleep feature.
This repository contains the source code of the SEN-DAL.
We evaluate our model on the Montreal Archive of Sleep Studies (MASS) dataset Subset-3. The MASS dataset is an open-access and collaborative database of laboratory-based polysomnography (PSG) recordings.
- Python 3.6.2
- CUDA 9.0
- CuDNN 7.6.4
- numpy==1.19.5
- sklearn==0.19.1
- tensorflow_gpu==1.8.0
- Keras==2.2.0
-
Preprocess
- Prepare data (raw signals) in
data_dir- Files name: 01-03-00XX-Data.npy, where XX denotes subject ID.
- Tensor shape: [sample, channel=26, length], where channel 0 is ECG channel, 1-20 are EEG channels, 21-23 are EMG channels, 24-25 are EOG channels.
- Modify directory
- Modify
data_dirandlabel_dirinrun_preprocess.pyaccordingly. - (Optional) Modify
output_dirinrun_preprocess.py.
- Modify
- Run preprocess program
cd preprocesspython run_preprocess.py
- Prepare data (raw signals) in
-
Training Command Line Arguments
- Training
--batch_size: Training batch size.--epoch: Number of training epochs.--num_fold: Number of folds.--save_model: Save the best model or not.--early_stop: Use early stop trick or not.--gpunums: Number of GPU(s).--from_fold: Training start fold.--to_fold: Training end fold.
- Directory
--label_dir: The directory of labels.--data_dir_eog: The directory of the EOG signals.--data_dir_psd: The directory of the spectral-spatial representation of EEG signals.--output: The directory for saving results.--model_dir: The directory for saving best models of each fold.
- Training
-
Training
Run
run.pywith the command line arguments. By default, the model can be run with the following command:CUDA_VISIBLE_DEVICES=0 python run.py