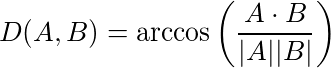K-means implementation is based on "Yinyang K-Means: A Drop-In Replacement of the Classic K-Means with Consistent Speedup". While it introduces some overhead and many conditional clauses which are bad for CUDA, it still shows 1.6-2x speedup against the Lloyd algorithm. K-nearest neighbors employ the same triangle inequality idea and require precalculated centroids and cluster assignments, similar to the flattened ball tree.
| Benchmarks | sklearn KMeans | KMeansRex | KMeansRex OpenMP | Serban | kmcuda | kmcuda 2 GPUs |
|---|---|---|---|---|---|---|
| speed | 1x | 4.5x | 8.2x | 15.5x | 17.8x | 29.8x |
| memory | 1x | 2x | 2x | 0.6x | 0.6x | 0.6x |
Technically, this project is a shared library which exports two functions
defined in kmcuda.h: kmeans_cuda and knn_cuda.
It has built-in Python3 and R native extension support, so you can
from libKMCUDA import kmeans_cuda or dyn.load("libKMCUDA.so").
- K-means
- K-nn
- Notes
- Building
- Testing
- Benchmarks
- Python examples
- Python API
- R examples
- R API
- C examples
- C API
- License
The major difference between this project and others is that kmcuda is optimized for low memory consumption and the large number of clusters. E.g., kmcuda can sort 4M samples in 480 dimensions into 40000 clusters (if you have several days and 12 GB of GPU memory); 300K samples are grouped into 5000 clusters in 4½ minutes on NVIDIA Titan X (15 iterations); 3M samples and 1000 clusters take 20 minutes (33 iterations). Yinyang can be turned off to save GPU memory but the slower Lloyd will be used then. Four centroid initialization schemes are supported: random, k-means++, AFKMC2 and import. Two distance metrics are supported: L2 (the usual one) and angular (arccos of the scalar product). L1 is in development. 16-bit float support delivers 2x memory compression. If you've got several GPUs, they can be utilized together and it gives the corresponding linear speedup either for Lloyd or Yinyang.
The code has been thoroughly tested to yield bit-to-bit identical results from Yinyang and Lloyd. "Fast and Provably Good Seedings for k-Means" was adapted from the reference code.
Centroid distance matrix Cij is calculated together with clusters' radiuses Ri (the maximum distance from the centroid to the corresponding cluster's members). Given sample S in cluster A, we avoid calculating the distances from S to another cluster B's members if CAB - SA - RB is greater than the current maximum K-nn distance. This resembles the ball tree algorithm.
The implemented algorithm is tolerant to NANs. There are two variants depending on whether k is small enough to fit the sample's neighbors into CUDA shared memory. Internally, the neighbors list is a binary heap - that reduces the complexity multiplier from O(k) to O(log k).
The implementation yields identical results to sklearn.neighbors.NearestNeighbors
except cases in which adjacent distances are equal and the order is undefined.
That is, the returned indices are sorted in the increasing order of the
corresponding distances.
Lloyd is tolerant to samples with NaN features while Yinyang is not. It may happen that some of the resulting clusters contain zero elements. In such cases, their features are set to NaN.
Angular (cosine) distance metric effectively results in Spherical K-Means behavior. The samples must be normalized to L2 norm equal to 1 before clustering, it is not done automatically. The actual formula is:
If you get OOM with the default parameters, set yinyang_t to 0 which
forces Lloyd. verbosity 2 will print the memory allocation statistics
(all GPU allocation happens at startup).
Data type is either 32- or 16-bit float. Number of samples is limited by 2^32, clusters by 2^32 and features by 2^16 (2^17 for fp16). Besides, the product of clusters number and features number may not exceed 2^32.
In the case of 16-bit floats, the reduced precision often leads to a slightly increased number of iterations, Yinyang is especially sensitive to that. In some cases, there may be overflows and the clustering may fail completely.
git clone https://github.com/src-d/kmcuda
cd src
cmake -DCMAKE_BUILD_TYPE=Release . && make
It requires cudart 8.0 / Pascal and OpenMP 4.0 capable compiler. The build has
been tested primarily on Linux but it works on macOS too with some blows and whistles
(see "macOS" subsection).
If you do not want to build the Python native module, add -D DISABLE_PYTHON=y.
If you do not want to build the R native module, add -D DISABLE_R=y.
If CUDA is not automatically found, add -D CUDA_TOOLKIT_ROOT_DIR=/usr/local/cuda-8.0
(change the path to the actual one). By default, CUDA kernels are compiled for
the architecture 60 (Pascal). It is possible to override it via -D CUDA_ARCH=52,
but fp16 support will be disabled then.
Python users:
CUDA_ARCH=61 pip install libKMCUDA
# replace 61 with your device version
Or install it from source:
CUDA_ARCH=61 pip install git+https://github.com/src-d/kmcuda.git#subdirectory=src
# replace 61 with your device version
Binary Python packages are quite hard to provide because they depend on CUDA and device architecture versions. PRs welcome!
macOS build is tricky, but possible. The instructions below correspond to the state from 1 year ago and may be different now. Please help with updates!
Install Homebrew and the Command Line Developer Tools
which are compatible with your CUDA installation. E.g., CUDA 8.0 does not support
the latest 8.x and works with 7.3.1 and below. Install clang with OpenMP support
and Python with numpy:
brew install llvm --with-clang
brew install python3
pip3 install numpy
Execute this magic command which builds kmcuda afterwards:
CC=/usr/local/opt/llvm/bin/clang CXX=/usr/local/opt/llvm/bin/clang++ LDFLAGS=-L/usr/local/opt/llvm/lib/ cmake -DCMAKE_BUILD_TYPE=Release .
And make the last important step - rename *.dylib to *.so so that Python is able to import the native extension:
mv libKMCUDA.{dylib,so}
test.py contains the unit tests based on unittest.
They require either cuda4py or pycuda and
scikit-learn.
test.R contains R integration tests and shall be run with Rscript.
| sklearn KMeans | KMeansRex | KMeansRex OpenMP | Serban | kmcuda | kmcuda 2 GPUs | |
|---|---|---|---|---|---|---|
| time, s | 164 | 36 | 20 | 10.6 | 9.2 | 5.5 |
| memory, GB | 1 | 2 | 2 | 0.6 | 0.6 | 0.6 |
- 16-core (32 threads) Intel Xeon E5-2620 v4 @ 2.10GHz
- 256 GB RAM Samsung M393A2K40BB1
- Nvidia Titan X 2016
- sklearn.cluster.KMeans@0.18.1;
KMeans(n_clusters=1024, init="random", max_iter=15, random_state=0, n_jobs=1, n_init=1). - KMeansRex@288c40a with
-march-nativeand Eigen 3.3;KMeansRex.RunKMeans(data, 1024, Niter=15, initname=b"random"). - KMeansRex with additional
-fopenmp. - Serban KMeans@83e76bf built for arch 6.1;
./cuda_main -b -i serban.bin -n 1024 -t 0.0028 -o - kmcuda v6.1 built for arch 6.1;
libKMCUDA.kmeans_cuda(dataset, 1024, tolerance=0.002, seed=777, init="random", verbosity=2, yinyang_t=0, device=0) - kmcuda running on 2 GPUs.
100000 random samples uniformly distributed between 0 and 1 in 256 dimensions.
100000 is the maximum size Serban KMeans can handle.
| sklearn KMeans | KMeansRex | KMeansRex OpenMP | Serban | kmcuda 2 GPU | kmcuda Yinyang 2 GPUs | |
|---|---|---|---|---|---|---|
| time | please no | - | 6h 34m | fail | 44m | 36m |
| memory, GB | - | - | 205 | fail | 8.7 | 10.4 |
kmeans++ initialization, 93 iterations (1% reassignments equivalent).
8,000,000 secret production samples.
KmeansRex did eat 205 GB of RAM on peak; it uses dynamic memory so it constantly bounced from 100 GB to 200 GB.
...are welcome! See CONTRIBUTING and code of conduct.
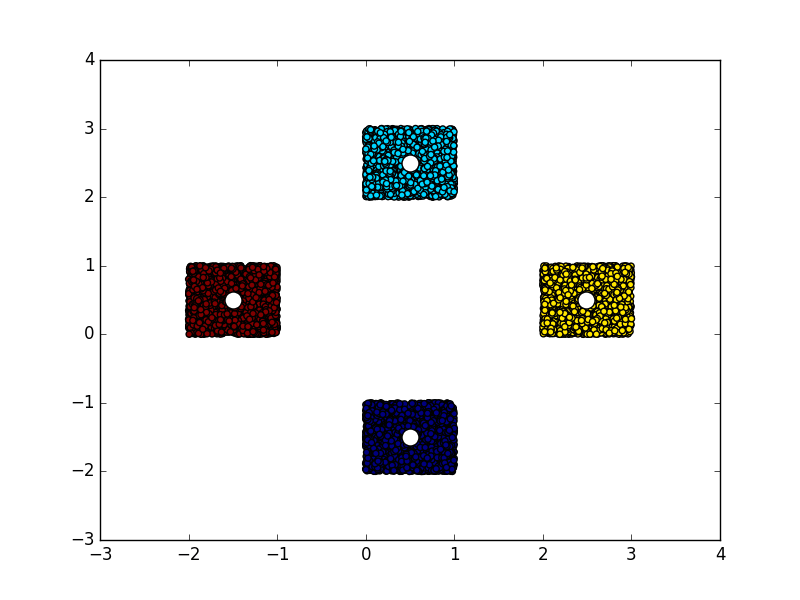
import numpy
from matplotlib import pyplot
from libKMCUDA import kmeans_cuda
numpy.random.seed(0)
arr = numpy.empty((10000, 2), dtype=numpy.float32)
arr[:2500] = numpy.random.rand(2500, 2) + [0, 2]
arr[2500:5000] = numpy.random.rand(2500, 2) - [0, 2]
arr[5000:7500] = numpy.random.rand(2500, 2) + [2, 0]
arr[7500:] = numpy.random.rand(2500, 2) - [2, 0]
centroids, assignments = kmeans_cuda(arr, 4, verbosity=1, seed=3)
print(centroids)
pyplot.scatter(arr[:, 0], arr[:, 1], c=assignments)
pyplot.scatter(centroids[:, 0], centroids[:, 1], c="white", s=150)You should see something like this:
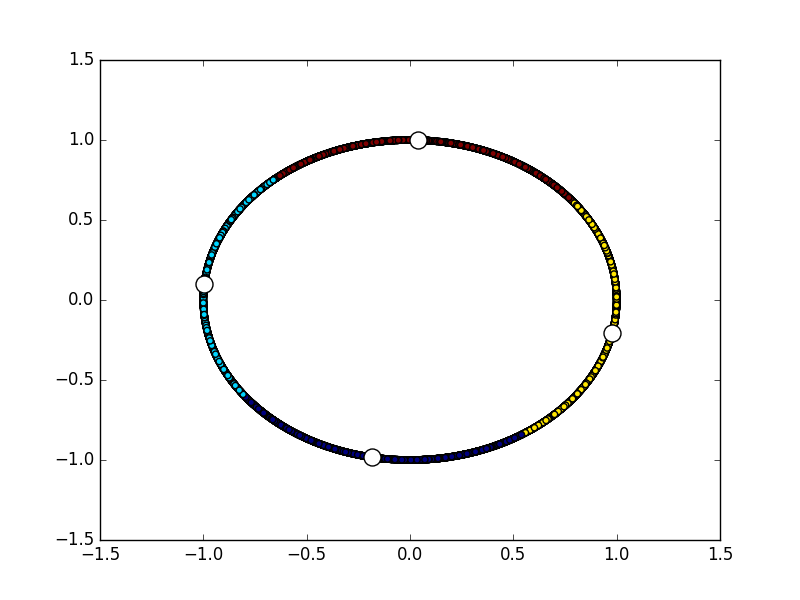
import numpy
from matplotlib import pyplot
from libKMCUDA import kmeans_cuda
numpy.random.seed(0)
arr = numpy.empty((10000, 2), dtype=numpy.float32)
angs = numpy.random.rand(10000) * 2 * numpy.pi
for i in range(10000):
arr[i] = numpy.sin(angs[i]), numpy.cos(angs[i])
centroids, assignments, avg_distance = kmeans_cuda(
arr, 4, metric="cos", verbosity=1, seed=3, average_distance=True)
print("Average distance between centroids and members:", avg_distance)
print(centroids)
pyplot.scatter(arr[:, 0], arr[:, 1], c=assignments)
pyplot.scatter(centroids[:, 0], centroids[:, 1], c="white", s=150)You should see something like this:
import numpy
from libKMCUDA import kmeans_cuda, knn_cuda
numpy.random.seed(0)
arr = numpy.empty((10000, 2), dtype=numpy.float32)
angs = numpy.random.rand(10000) * 2 * numpy.pi
for i in range(10000):
arr[i] = numpy.sin(angs[i]), numpy.cos(angs[i])
ca = kmeans_cuda(arr, 4, metric="cos", verbosity=1, seed=3)
neighbors = knn_cuda(10, arr, *ca, metric="cos", verbosity=1, device=1)
print(neighbors[0])You should see
reassignments threshold: 100
performing kmeans++...
done
too few clusters for this yinyang_t => Lloyd
iteration 1: 10000 reassignments
iteration 2: 926 reassignments
iteration 3: 416 reassignments
iteration 4: 187 reassignments
iteration 5: 87 reassignments
initializing the inverse assignments...
calculating the cluster radiuses...
calculating the centroid distance matrix...
searching for the nearest neighbors...
calculated 0.276552 of all the distances
[1279 1206 9846 9886 9412 9823 7019 7075 6453 8933]
def kmeans_cuda(samples, clusters, tolerance=0.01, init="k-means++",
yinyang_t=0.1, metric="L2", average_distance=False,
seed=time(), device=0, verbosity=0)samples numpy array of shape [number of samples, number of features] or tuple(raw device pointer (int), device index (int), shape (tuple(number of samples, number of features[, fp16x2 marker]))). In the latter case, negative device index means host pointer. Optionally, the tuple can be 2 items longer with preallocated device pointers for centroids and assignments. dtype must be either float16 or convertible to float32.
clusters integer, the number of clusters.
tolerance float, if the relative number of reassignments drops below this value, algorithm stops.
init string or numpy array, sets the method for centroids initialization, may be "k-means++", "afk-mc2", "random" or numpy array of shape [clusters, number of features]. dtype must be float32.
yinyang_t float, the relative number of cluster groups, usually 0.1. 0 disables Yinyang refinement.
metric str, the name of the distance metric to use. The default is Euclidean (L2), it can be changed to "cos" to change the algorithm to Spherical K-means with the angular distance. Please note that samples must be normalized in the latter case.
average_distance boolean, the value indicating whether to calculate the average distance between cluster elements and the corresponding centroids. Useful for finding the best K. Returned as the third tuple element.
seed integer, random generator seed for reproducible results.
device integer, bitwise OR-ed CUDA device indices, e.g. 1 means first device, 2 means second device, 3 means using first and second device. Special value 0 enables all available devices. The default is 0.
verbosity integer, 0 means complete silence, 1 means mere progress logging, 2 means lots of output.
return tuple(centroids, assignments, [average_distance]). If samples was a numpy array or a host pointer tuple, the types are numpy arrays, otherwise, raw pointers (integers) allocated on the same device. If samples are float16, the returned centroids are float16 too.
def knn_cuda(k, samples, centroids, assignments, metric="L2", device=0, verbosity=0)k integer, the number of neighbors to search for each sample. Must be ≤ 116.
samples numpy array of shape [number of samples, number of features] or tuple(raw device pointer (int), device index (int), shape (tuple(number of samples, number of features[, fp16x2 marker]))). In the latter case, negative device index means host pointer. Optionally, the tuple can be 1 item longer with the preallocated device pointer for neighbors. dtype must be either float16 or convertible to float32.
centroids numpy array with precalculated clusters' centroids (e.g., using K-means/kmcuda/kmeans_cuda()). dtype must match samples. If samples is a tuple then centroids must be a length-2 tuple, the first element is the pointer and the second is the number of clusters. The shape is (number of clusters, number of features).
assignments numpy array with sample-cluster associations. dtype is expected to be compatible with uint32. If samples is a tuple then assignments is a pointer. The shape is (number of samples,).
metric str, the name of the distance metric to use. The default is Euclidean (L2), it can be changed to "cos" to change the algorithm to Spherical K-means with the angular distance. Please note that samples must be normalized in the latter case.
device integer, bitwise OR-ed CUDA device indices, e.g. 1 means first device, 2 means second device, 3 means using first and second device. Special value 0 enables all available devices. The default is 0.
verbosity integer, 0 means complete silence, 1 means mere progress logging, 2 means lots of output.
return neighbor indices. If samples was a numpy array or a host pointer tuple, the return type is numpy array, otherwise, a raw pointer (integer) allocated on the same device. The shape is (number of samples, k).
dyn.load("libKMCUDA.so")
samples = replicate(4, runif(16000))
result = .External("kmeans_cuda", samples, 50, tolerance=0.01,
seed=777, verbosity=1, average_distance=TRUE)
print(result$average_distance)
print(result$centroids[1:10,])
print(result$assignments[1:10])dyn.load("libKMCUDA.so")
samples = replicate(4, runif(16000))
cls = .External("kmeans_cuda", samples, 50, tolerance=0.01,
seed=777, verbosity=1)
result = .External("knn_cuda", 20, samples, cls$centroids, cls$assignments,
verbosity=1)
print(result[1:10,])function kmeans_cuda(
samples, clusters, tolerance=0.01, init="k-means++", yinyang_t=0.1,
metric="L2", average_distance=FALSE, seed=Sys.time(), device=0, verbosity=0)samples real matrix of shape [number of samples, number of features] or list of real matrices which are rbind()-ed internally. No more than INT32_MAX samples and UINT16_MAX features are supported.
clusters integer, the number of clusters.
tolerance real, if the relative number of reassignments drops below this value, algorithm stops.
init character vector or real matrix, sets the method for centroids initialization, may be "k-means++", "afk-mc2", "random" or real matrix, of shape [clusters, number of features].
yinyang_t real, the relative number of cluster groups, usually 0.1. 0 disables Yinyang refinement.
metric character vector, the name of the distance metric to use. The default is Euclidean (L2), it can be changed to "cos" to change the algorithm to Spherical K-means with the angular distance. Please note that samples must be normalized in the latter case.
average_distance logical, the value indicating whether to calculate the average distance between cluster elements and the corresponding centroids. Useful for finding the best K. Returned as the third list element.
seed integer, random generator seed for reproducible results.
device integer, bitwise OR-ed CUDA device indices, e.g. 1 means first device, 2 means second device, 3 means using first and second device. Special value 0 enables all available devices. The default is 0.
verbosity integer, 0 means complete silence, 1 means mere progress logging, 2 means lots of output.
return list(centroids, assignments[, average_distance]). Indices in assignments start from 1.
function knn_cuda(k, samples, centroids, assignments, metric="L2", device=0, verbosity=0)k integer, the number of neighbors to search for each sample. Must be ≤ 116.
samples real matrix of shape [number of samples, number of features] or list of real matrices which are rbind()-ed internally. In the latter case, is is possible to pass in more than INT32_MAX samples.
centroids real matrix with precalculated clusters' centroids (e.g., using kmeans() or kmeans_cuda()).
assignments integer vector with sample-cluster associations. Indices start from 1.
metric str, the name of the distance metric to use. The default is Euclidean (L2), can be changed to "cos" to behave as Spherical K-means with the angular distance. Please note that samples must be normalized in that case.
device integer, bitwise OR-ed CUDA device indices, e.g. 1 means first device, 2 means second device, 3 means using first and second device. Special value 0 enables all available devices. The default is 0.
verbosity integer, 0 means complete silence, 1 means mere progress logging, 2 means lots of output.
return integer matrix with neighbor indices. The shape is (number of samples, k). Indices start from 1.
example.c:
#include <assert.h>
#include <stdint.h>
#include <stdio.h>
#include <stdlib.h>
#include <kmcuda.h>
// ./example /path/to/data <number of clusters>
int main(int argc, const char **argv) {
assert(argc == 3);
// we open the binary file with the data
// [samples_size][features_size][samples_size x features_size]
FILE *fin = fopen(argv[1], "rb");
assert(fin);
uint32_t samples_size, features_size;
assert(fread(&samples_size, sizeof(samples_size), 1, fin) == 1);
assert(fread(&features_size, sizeof(features_size), 1, fin) == 1);
uint64_t total_size = ((uint64_t)samples_size) * features_size;
float *samples = malloc(total_size * sizeof(float));
assert(samples);
assert(fread(samples, sizeof(float), total_size, fin) == total_size);
fclose(fin);
int clusters_size = atoi(argv[2]);
// we will store cluster centers here
float *centroids = malloc(clusters_size * features_size * sizeof(float));
assert(centroids);
// we will store assignments of every sample here
uint32_t *assignments = malloc(((uint64_t)samples_size) * sizeof(uint32_t));
assert(assignments);
float average_distance;
KMCUDAResult result = kmeans_cuda(
kmcudaInitMethodPlusPlus, NULL, // kmeans++ centroids initialization
0.01, // less than 1% of the samples are reassigned in the end
0.1, // activate Yinyang refinement with 0.1 threshold
kmcudaDistanceMetricL2, // Euclidean distance
samples_size, features_size, clusters_size,
0xDEADBEEF, // random generator seed
0, // use all available CUDA devices
-1, // samples are supplied from host
0, // not in float16x2 mode
1, // moderate verbosity
samples, centroids, assignments, &average_distance);
free(samples);
free(centroids);
free(assignments);
assert(result == kmcudaSuccess);
printf("Average distance between a centroid and the corresponding "
"cluster members: %f\n", average_distance);
return 0;
}Build:
gcc -std=c99 -O2 example.c -I/path/to/kmcuda.h/dir -L/path/to/libKMCUDA.so/dir -l KMCUDA -Wl,-rpath,. -o example
Run:
./example serban.bin 1024
The file format is the same as in serban/kmeans.
KMCUDAResult kmeans_cuda(
KMCUDAInitMethod init, float tolerance, float yinyang_t,
KMCUDADistanceMetric metric, uint32_t samples_size, uint16_t features_size,
uint32_t clusters_size, uint32_t seed, uint32_t device, int32_t device_ptrs,
int32_t fp16x2, int32_t verbosity, const float *samples, float *centroids,
uint32_t *assignments, float *average_distance)init specifies the centroids initialization method: k-means++, random or import (in the latter case, centroids is read).
tolerance if the number of reassignments drop below this ratio, stop.
yinyang_t the relative number of cluster groups, usually 0.1.
metric The distance metric to use. The default is Euclidean (L2), can be changed to cosine to behave as Spherical K-means with the angular distance. Please note that samples must be normalized in that case.
samples_size number of samples.
features_size number of features. if fp16x2 is set, one half of the number of features.
clusters_size number of clusters.
seed random generator seed passed to srand().
device CUDA device OR-ed indices - usually 1. For example, 1 means using first device, 2 means second device, 3 means first and second device (2x speedup). Special value 0 enables all available devices.
device_ptrs configures the location of input and output. If it is negative, samples and returned arrays are on host, otherwise, they belong to the corresponding device. E.g., if device_ptrs is 0, samples is expected to be a pointer to device #0's memory and the resulting centroids and assignments are expected to be preallocated on device #0 as well. Usually this value is -1.
fp16x2 activates fp16 mode, two half-floats are packed into a single 32-bit float, features_size becomes effectively 2 times bigger, the returned centroids are fp16x2 too.
verbosity 0 - no output; 1 - progress output; >=2 - debug output.
samples input array of size samples_size x features_size in row major format.
centroids output array of centroids of size clusters_size x features_size in row major format.
assignments output array of cluster indices for each sample of size samples_size x 1.
average_distance output mean distance between cluster elements and the corresponding centroids. If nullptr, not calculated.
Returns KMCUDAResult (see kmcuda.h);
KMCUDAResult knn_cuda(
uint16_t k, KMCUDADistanceMetric metric, uint32_t samples_size,
uint16_t features_size, uint32_t clusters_size, uint32_t device,
int32_t device_ptrs, int32_t fp16x2, int32_t verbosity,
const float *samples, const float *centroids, const uint32_t *assignments,
uint32_t *neighbors);k integer, the number of neighbors to search for each sample.
metric The distance metric to use. The default is Euclidean (L2), can be changed to cosine to behave as Spherical K-means with the angular distance. Please note that samples must be normalized in that case.
samples_size number of samples.
features_size number of features. if fp16x2 is set, one half of the number of features.
clusters_size number of clusters.
device CUDA device OR-ed indices - usually 1. For example, 1 means using first device, 2 means second device, 3 means first and second device (2x speedup). Special value 0 enables all available devices.
device_ptrs configures the location of input and output. If it is negative, samples, centroids, assignments and the returned array are on host, otherwise, they belong to the corresponding device. E.g., if device_ptrs is 0, samples, centroids and assignments are expected to be pointers to device #0's memory and the resulting neighbors is expected to be preallocated on device #0 as well. Usually this value is -1.
fp16x2 activates fp16 mode, two half-floats are packed into a single 32-bit float, features_size becomes effectively 2 times bigger, affects samples and centroids.
verbosity 0 - no output; 1 - progress output; >=2 - debug output.
samples input array of size samples_size x features_size in row major format.
centroids input array of centroids of size clusters_size x features_size in row major format.
assignments input array of cluster indices for each sample of size samples_size x 1.
neighbors output array with the nearest neighbors of size samples_size x k in row major format.
Returns KMCUDAResult (see kmcuda.h);