A toolkit for haplotype analysis using haplotype sharing trees, haplotype windowing and long-read assemblied haplotypes
# Install Rust and the Cargo package manager
curl --proto '=https' --tlsv1.2 -sSf https://sh.rustup.rs | sh
# The Rust binaries directory is added automatically to the shells $PATH variable when it is restarted
# If you face problems, add the directory to the $PATH variable.
# For example for `bash` or `zsh`, run this line:
export PATH="$HOME/.cargo/bin:$PATH"
# For `fish` shell, use this command:
fish_add_path "$HOME/.cargo/bin"
# Make sure cmake is installed
# It is available in all common package managers such as apt, dnf, brew
# Install HAPTK using cargo (recommended), or download a binary from the releases page
cargo install haptk --locked
# Install the python graph library using anaconda or micromamba (environment.yml is located at the github root directory)
micromamba env create -f environment.yml
micromamba activate haptk
# OR by running pip. Using python 3.12 is highly recommended.
pip install haptkA common requirement is to select only certain alleles of the samples based on a condition, this is done with the --alleles argument.
Options:
all(default) [Select all alleles (diploid genomes)]only-refs[Select only the samples carrying the REF variant at the coordinate]only-alts[Select only the samples carrying the ALT variant at the coordinate]longest-haplotype[Select only the allele per each sample sharing the most haplotype with the haplotypes of the other samples]
# Go into the examples directory
cd examples
# Download the 1000 genomes reference panel for chromosome 9
wget ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20220422_3202_phased_SNV_INDEL_SV/1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.vcf.gz
wget ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20220422_3202_phased_SNV_INDEL_SV/1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.vcf.gz.tbi
# Extract biallelic SNPs from the panel
bcftools view -m2 -M2 -v snps 1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.vcf.gz -Ou |
bcftools view -R data/illumina_gsa_v3_hg38.tsv.gz -Ou | # To ease the computational load, you can select variants from an SNP array
bcftools annotate -x "INFO" -Oz -o 1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.biallelic.vcf.gz
# Index the file
bcftools index -t 1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.biallelic.vcf.gz
# Create unidirectional HSTs of the 1kg samples
haptk uhst 1kGP_high_coverage_Illumina.chr9.filtered.SNV_INDEL_SV_phased_panel.biallelic.vcf.gz \
--alleles all \
--samples 1kGP_high_coverage_Illumina.finnish.ids 1kGP_high_coverage_Illumina.gambian.ids 1kGP_high_coverage_Illumina.han_chinese.ids \
--coords chr9:27573534 \
-t 8 \
-o results
Visualize the HST using the HAPTK Python library
import haptk
def read_samples(filename):
with open(filename) as file:
lines = [line.rstrip() for line in file]
return lines
# The IDs are located in the examples directory
gambian = read_samples("1kGP_high_coverage_Illumina.gambian.ids")
finnish = read_samples("1kGP_high_coverage_Illumina.finnish.ids")
han_chinese = read_samples("1kGP_high_coverage_Illumina.han_chinese.ids")
hst = haptk.read_hst("results/uhst_left.hst.gz")
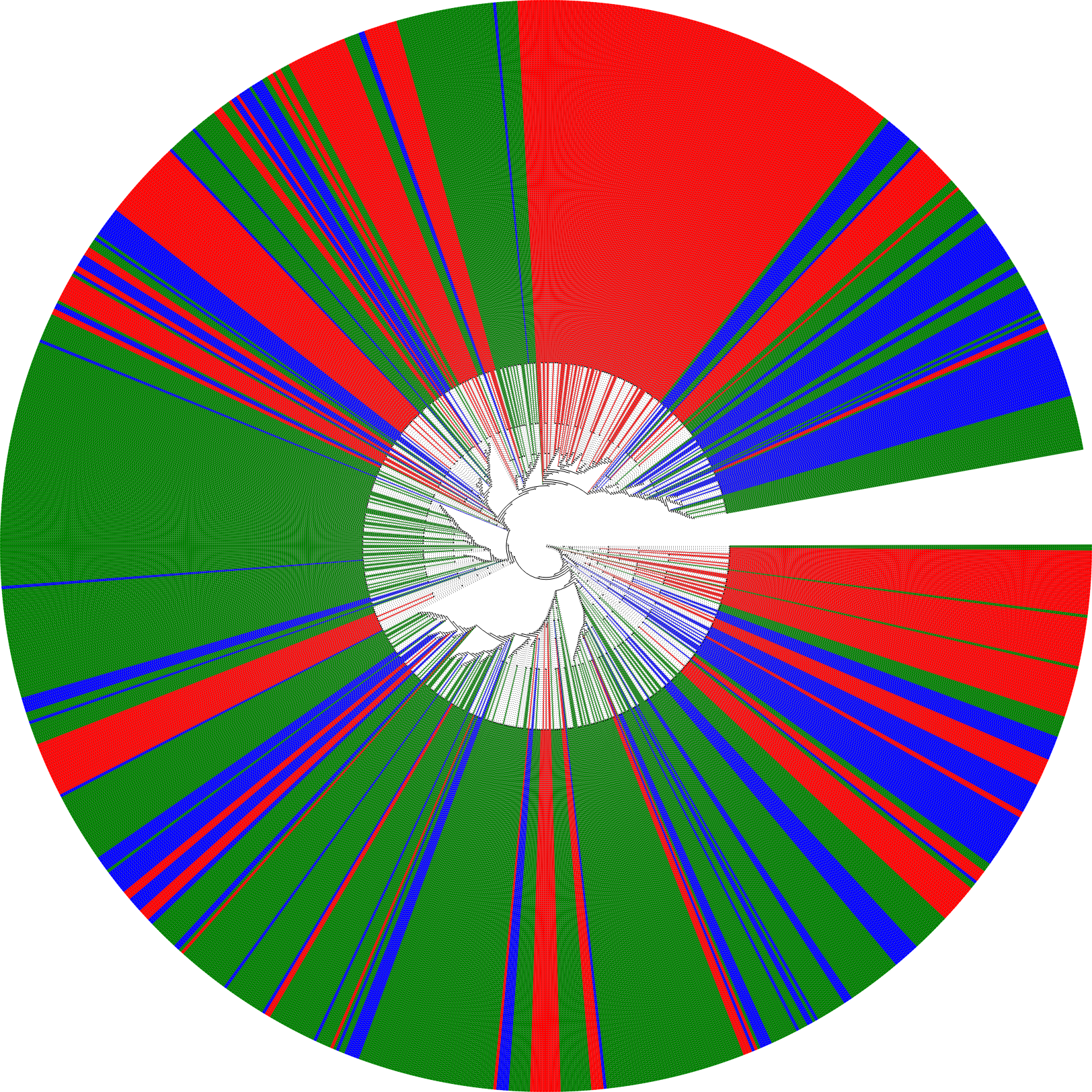
hst.circle_tree("my_left_hst.png", to_tag=[gambian, finnish, han_chinese], colors=["red", "blue", "green"])The HST starting from chr9:27573534 tagged for Gambian (red), Finnish (blue) and Han Chinese (green) ancestry
Commands:
uhst Build unidirectional haplotype sharing trees at a coordinate
bhst Build a bidirectional haplotype sharing tree at a coordinate
mrca Analyze the MRCA based on the Gamma method at a coordinate
check-for-haplotype Check if samples share a given haplotype
compare-to-haplotype Check differences between samples and a haplotype
compare-to-hst Check what haplotypes of the HST are present in samples
compare-haplotypes Compare haplotypes to each other by alignment
haplotypes Read the haplotypes of a given sample
samples Output the sample names from FAM / VCF / HST files
markers Output the markers from a HST file
haplotype-to-vcf Convert a haplotype CSV into VCF
fasta-to-haplotype Convert fasta sequences to haplotype csv format
help Print this message or the help of the given subcommand(s)
Example graphs from the original article
The left side uHST starting from the C9orf72 expansion.
The right side uHST starting from the C9orf72 expansion.
A Finnish C9orf72 expansion carrier bHST with FTD cases tagged in green and ALS cases tagged in red.
All the samples are compared to a haplotype, in this case the C9orf72 expansion ancestral haplotype. The matching variants are colored in white (or blue) and deviating variants in pink. Because the actual sample haplotypes are seen in relation to the ancestral haplotype, potential switch and genotyping errors can be evaluated from this graph. The selection of C9orf72 haplotypes is done by discarding the haplotypes tagged in blue (the ones sharing less ancestral haplotype sequence per sample).
The shared ancestry between the C9orf72 hexanucleotide repeat expansion and intermediate-length alleles using haplotype sharing trees and HAPTK
https://pubmed.ncbi.nlm.nih.gov/38242117/
@article{rautila2024shared,
title={The shared ancestry between the C9orf72 hexanucleotide repeat expansion and intermediate-length alleles using haplotype sharing trees and HAPTK},
author={Rautila, Osma S and Kaivola, Karri and Rautila, Harri and Hokkanen, Laura and Launes, Jyrki and Strandberg, Timo E and Laaksovirta, Hannu and Palmio, Johanna and Tienari, Pentti J},
journal={The American Journal of Human Genetics},
volume={111},
number={2},
pages={383--392},
year={2024},
publisher={Elsevier}
}
# Clone the repository
git clone https://github.com/xosxos/haptk
# Change directory
cd haptk
# Build. The binary will be available in `./target/release/`
cargo build --release --bin haptk