This is our implementation for the paper:
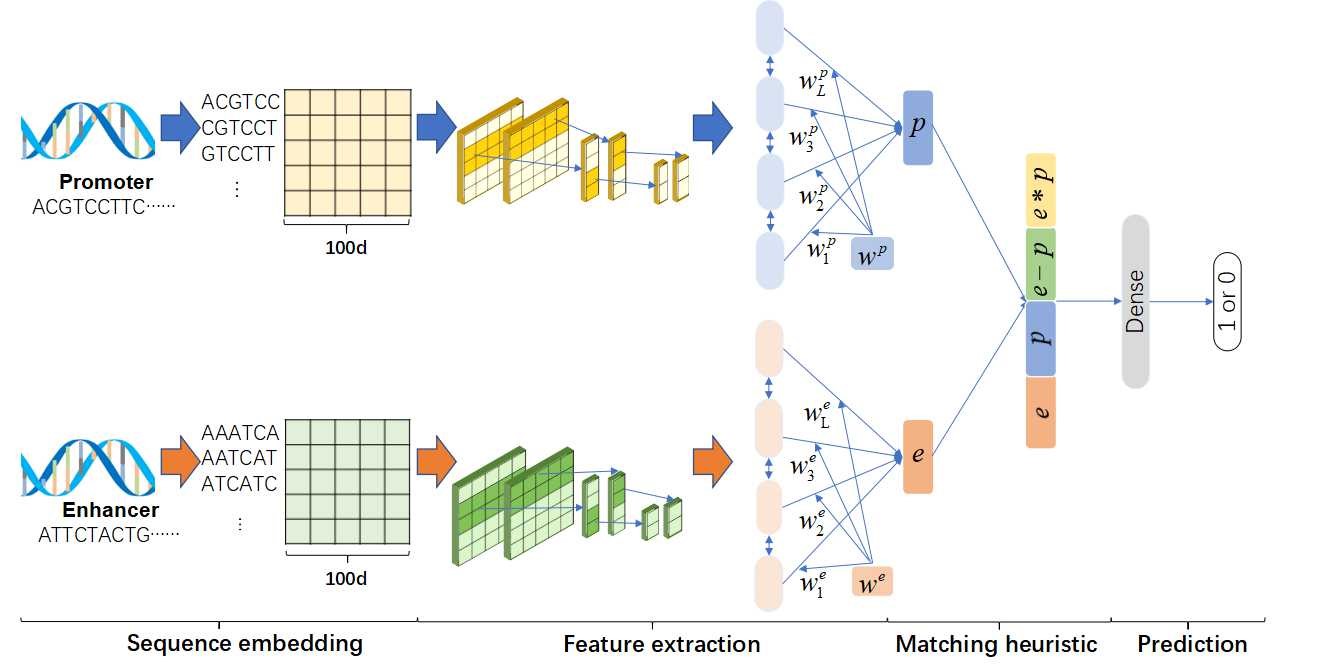
Figure1 shows the Architecture of the model. It consists of four steps, including Sequence embedding, Feature extraction, Matching heuristic, and Prediction.Predicting Enhancer-Promoter Interactions by Deep Learning and Matching Heuristic
Due to size limitation, we put the paper data on Zenodo, available from https://zenodo.org/record/4018229
-
Data_Augmentation.R
A tool of data augmentation provided by Mao et al. (2017). The details of the tool can be seen in https://github.com/wgmao/EPIANN.
-
sequence_processing.py
Used for pre-processing DNA sequences
-
embedding_matrix.npy
The weight of the embedding layer converted from the pre-trained DNA vector provided by Ng (2017).
-
train.py, train_c_-.py, train_c_x.py, train_max.py
Used for training all EPI-DLMH models
-
test.py
Evaluate the performance of models.
1.python sequence_processing.py
2.python train.py
3.python test.py