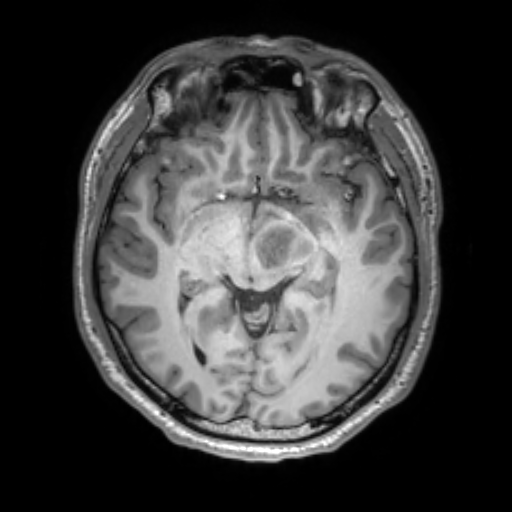
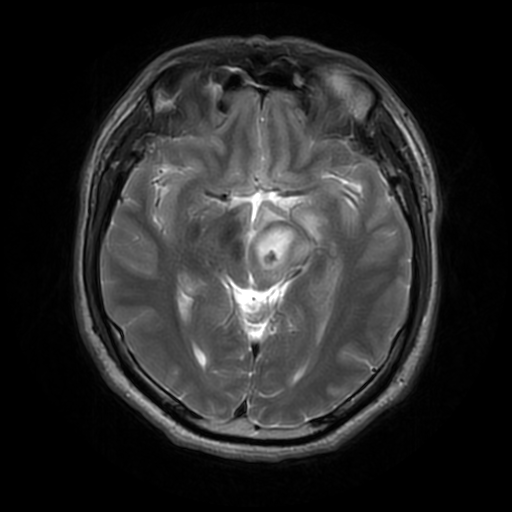
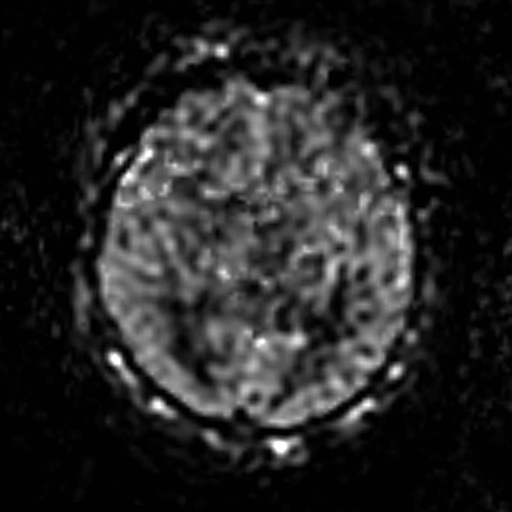
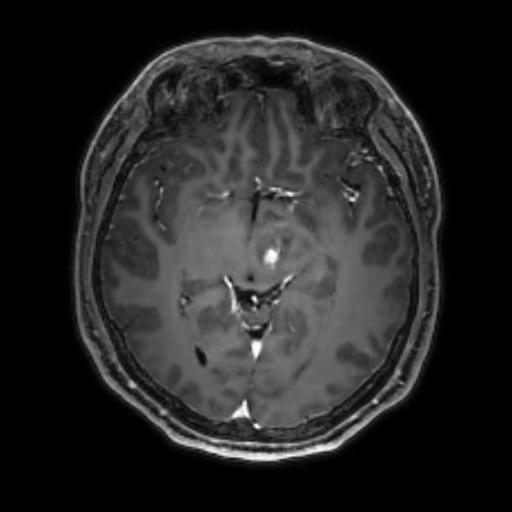
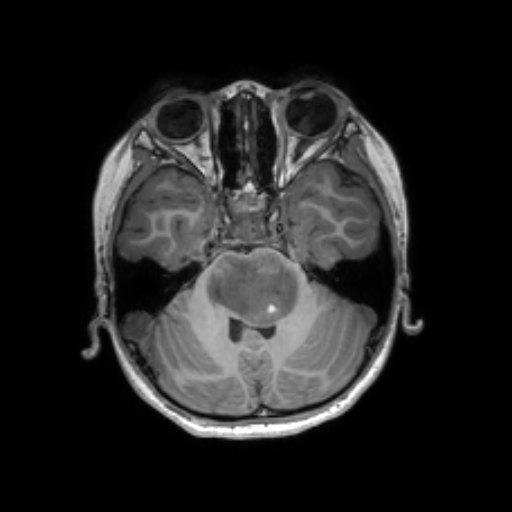
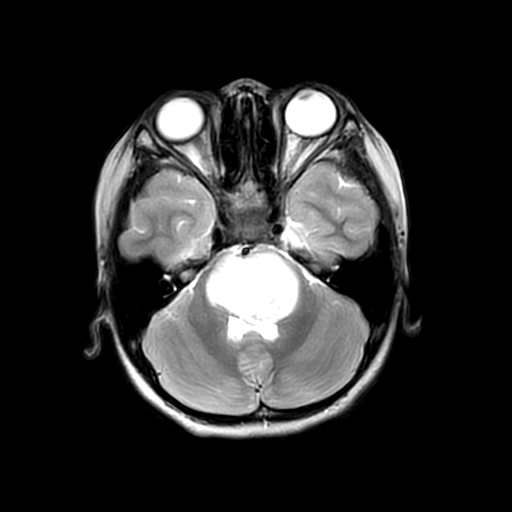
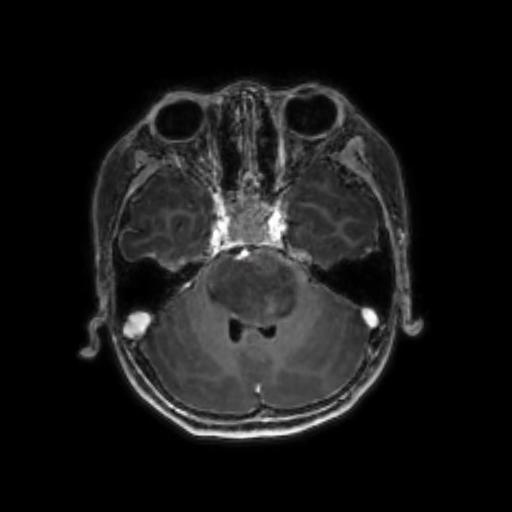
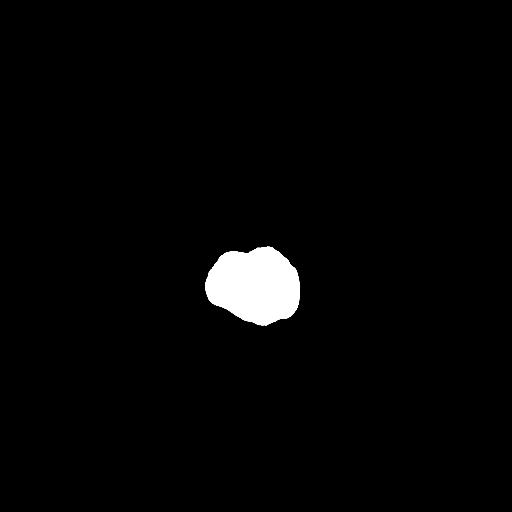
To reduce the exposure of Gadolinium-based Contrast Agents (GBCAs) in brainstem glioma detection and provide high-resolution contrast information,
we propose a novel multi-task generative network for contrast-enhanced T1-weight MR synthesis on brainstem glioma images. The proposed network
can simultaneously synthesize the high-resolution contrast-enhanced image and the segmentation mask of brainstem glioma lesions.
- {T1, T2, ASL}-to-{T1ce, tumor mask}
- Linux or Windows
- Python 3
- NVIDIA GPU (11G memory or larger) + CUDA cuDNN
- Install PyTorch and dependencies from http://pytorch.org
- Install python libraries dominate.
pip install dominate- Clone this repo:
git clone https://github.com/yXiangXiong/Multi-task_Generative_Synthesis_Network
cd pix2pixHD_Multi-task_Learning- Test the model (
bash ./scripts/test_1024p.sh):
#!./scripts/test.sh
python test.py --dataroot F:\xiongxiangyu\pix2pixHD_Mask_Data --name NC2C --label_nc 0 --input_nc 9 --output_nc 6 --resize_or_crop none --gpu_ids 0 --which_epoch 200 --no_instance --how_many 144The test results will be saved to a html file here: ./results/NC2C/test_latest/index.html.
More example scripts can be found in the scripts directory.
- Train a model at 512 x 512 resolution (
bash ./scripts/train_512p.sh):
#!./scripts/train.sh
python train.py --dataroot F:\xiongxiangyu\pix2pixHD_Mask_Data --name NC2C --label_nc 0 --input_nc 9 --output_nc 6 --netG global --resize_or_crop none --gpu_ids 0 --batchSize 1 --no_instance- To view training results, please checkout intermediate results in
./checkpoints/NC2C/web/index.html. If you have tensorflow installed, you can see tensorboard logs in./checkpoints/NC2C/logsby adding--tf_logto the training scripts.
If you find this useful for your research, please use the following.
This code borrows heavily from pix2pixHD.