This is a Python script used in Sakata, Ishiguro, Mori et al., Nat. Biotech. (2020) to predict frequencies of base editing patterns for a given input sequence using a model trained with amplicon sequencing data obtained for a specific base editing method. Let 





sample_training_codes/).
In this script, a predicted frequency of a given editing pattern for an input target sequence is calculated by the following formula:
where




This script also enables prediction of all possible base editing pattern frequencies for a given input target sequence and generates an expected editing spectrum with total base editing frequencies across different positions relative to the PAM.
Python 3.7.0 or later
1. Download the software by
git clone https://github.com/yachielab/base-editing-prediction
2. Install the necessary Python packages
pip install matplotlib
pip install numpy
SYNOPSIS
base-editing-prediction.py [-i] [-o] [-s] [-e] [-m] [-f][-h] [--help]
Options:
-i <String>
Input target sequence
-o <String>
Editing outcome sequence
-s <Integer>
Start position of the input target sequence relative to the PAM
-e <Integer>
End position of the input target sequence relative to the PAM
-m <String>
File path of base editing model
-f <String>
Output file path and name. Ex. [file_path]/[output]
-h, --help
Print the usage of this script
python base-editing-prediction.py -i ACACACACACACTCTGATCATACGA -o ACACACATGTGCTCTGATCATACGA -e '-1' -m sample_models/TargetACEmax.csv
Output : Standard output as follows
Model name : Target-ACEmax
Target sequence : ACACACACACACTCTGATCATACGA
Editing outcome : ACACACATGTGCTCTGATCATACGA
Editing frequency : 0.01109914561215845
Example 2: Predicting frequencies of all the possible base editing patterns for a given target sequence
python base-editing-prediction.py -i ACACACACTCTGATCATACGAGGG -s '-21' -m sample_models/TargetACEmax.csv -f [file_path]/[output]
Output : Generates the following three files in [file_path]
1. [output]_allpatterns.csv
A csv file showing all of the possible base editing patterns for a given target sequence and the frequency of their outcomes. The editing patterns are sorted by their frequencies.
#Model name : Target-ACEmax
#Target sequence : ACACACACTCTGATCATACGAGGG
#Start position : -21
#End poition : 2
Editing outcome,Editing frequency
ACATACGCTCTGATCATACGAGGG,0.030383595326181866
ACATATGCTCTGATCATACGAGGG,0.023406517399228513
ACATACACTCTGATCATACGAGGG,0.02268547605844774
ATATATGCTCTGATCATACGAGGG,0.014662449890585282
ACATGCGCTCTGATCATACGAGGG,0.014346124947638123
ATATACGCTCTGATCATACGAGGG,0.013568609108007466
ACATATACTCTGATCATACGAGGG,0.012644536102221833
ACATGTGCTCTGATCATACGAGGG,0.011103190756267084
ACATGCACTCTGATCATACGAGGG,0.01046274879400725
ATATGCGCTCTGATCATACGAGGG,0.008231783270293863
...
2. [output]_spectrum.csv
A csv file showing total frequencies of the three possible base transition patterns in every position across the target sequence.
#Model name : Target-ACEmax
#Target sequence : ACACACACTCTGATCATACGAGGG
#Start position : -21,
#End position : 2,
Position from the PAM,Target nucleotide,Frequency of A,Frequency of T,Frequency of G,Frequency of C
-21,A,0,0,0,0
-20,C,0.0006907111572225605,0.10090512721350689,0.0006542754524790089,0
-19,A,0,0.0,0.004490670388906142,0.0
-18,C,0.0026539014383369164,0.25930843007850085,0.006832040880274,0
-17,A,0,0.00011751821751747789,0.10173193279973115,1.8519962276380747e-05
-16,C,0.0010477675769179355,0.13061827086717823,0.0007392006189616716,0
-15,A,0,0.00012412091255604963,0.18688455821841132,3.6734772493093755e-05
-14,C,0.0002451145136467862,0.03804442863762912,0.0007554815817277042,0
-13,T,0,0,0,0
-12,C,0.00024972693713097945,0.024341927871551598,0.0009447857670925626,0
-11,T,0,0,0,0
-10,G,0,0,0,0
...
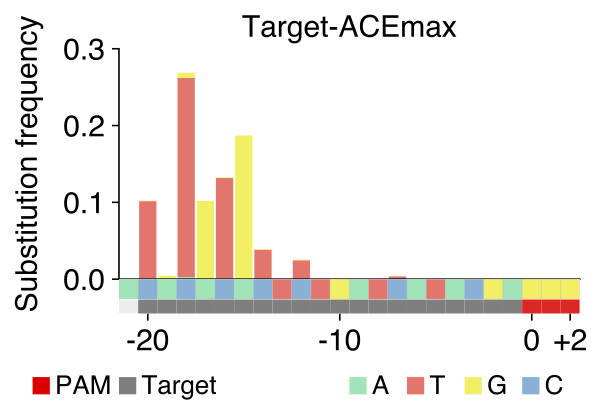
3.[output]_spectrum.pdf
A pdf file visualizing total frequencies of the three possible base transition patterns in every position across the target sequence.
A base editor model needs to be prepared as a csv file in the following format.
#Model Name : Target-ACEmax
#TP : Target transition probability
#CTP : Conditional transition probability
Data type,Conditional base transition,Target base transition,Probability
TP,,-30:A>A,0
TP,,-30:A>T,0
TP,,-30:A>G,1.0102950035855926e-05
TP,,-30:A>C,0
TP,,-30:T>A,1.986441510704398e-05
TP,,-30:T>T,0
TP,,-30:T>G,0
TP,,-30:T>C,3.1252658956434616e-05
TP,,-30:G>A,0
TP,,-30:G>T,0.000140296703177661
...
CTP,-30:A>A,-30:A>A,1.0
CTP,-30:A>A,-30:A>T,0
CTP,-30:A>A,-30:A>G,0
CTP,-30:A>A,-30:A>C,0
CTP,-30:A>T,-30:A>A,0
CTP,-30:A>T,-30:A>T,1.0
CTP,-30:A>T,-30:A>G,0
CTP,-30:A>T,-30:A>C,0
...
Data type: TP (transition probability) or CTP (Conditional transition probability)
Conditional base transition: [Relative position from the PAM]:[nucleotide transition pattern]. This should be left empty when the data type is TP, or ignored.
Target base transition: [Relative position from the PAM]:[nucleotide transition pattern]
Probability: Probability of the target base transition (TP) or probability of the target base transition given the conditional base transition (CTP).
Base editing models 13 different base editing methods used in Sakata, Ishiguro, Mori et al. (2020) are provided in sample_models/. All of the base editing models were created for a target sequence region from -30 bp to +10 bp to the PAM.
Cytosine base editors (CBEs):
sample_models/TargetAID.csv
sample_models/TargetAIDmax.csv
sample_models/BE4max.csv
sample_models/BE4maxC.csv
Adenine base editors (ABEs):
sample_models/ABE.csv
sample_models/ABEmax.csv
Base editor mixes:
sample_models/TargetAID_plus_ABE.csv
sample_models/TargetAIDmax_plus_ABEmax.csv
sample_models/BE4max_plus_ABEmax.csv
sample_models/BE4maxC_plus_ABEmax.csv
Dual function base editors
sample_models/TargetACE.csv
sample_models/TargetACEmax.csv
sample_models/ACBEmax.csv