- This code combines
emceeandRADEX, performing MCMC sampling in the RADEX-parameters space WITHOUT predefined grids, which leads to faster a convergence time and a better sampling of the parameter space. - This code directly samples the parameter space with the Affine Invariant Markov chain Monte Carlo (MCMC) Ensemble sampler. This allows a much better sampling of the parameter space.
This is a very valid concern. Nevertheless, it is always the first practice to test your code on well-studied sources (and you can also test the code yourself by comparing with literatures). That's what has been done:
- This code has been tested against a few well-studied sources. For example:
- APM08279+5255, Weiss et al. 2007: our results are entirely consistent with the results in this paper. Their values, log(n_H2/cm⁻³) = 4.2 and log(T_K/K) = 2.4, are within the +/-1 sigma values and close to the maximum likelihood values from the calculations based on this code.
- The Circinus Galaxy, Zhang et al. 2014: similar to APM08279+5255, the results from our code are fully consistent with the results in the reference.
PyRadex: https://github.com/keflavich/pyradexemcee: https://github.com/dfm/emceecorner: https://github.com/dfm/corner.py- other common packages:
numpy,scipy,astropy,astroquery - note that if you would like to directly use the compiled radex in this repo, you need to run the x86_64 version of python
- The results are stored in the
.picklefiles - Just run
replot(“source_name”)after executing the python script inIPython. - You can always check the
.picklefiles without the need to rerun the whole code again. - A note on a possible decoding error for the Pickle file (Python 2 --> Python 3)
- When you encounter an error using old Pickle file generated using Python 2:
UnicodeDecodeError: 'ascii' codec can't decode byte 0xfa in position 0: ordinal not in range(128) - You can add
, encoding='latin1'behind the line= pickle.load(pkl_file), so as= pickle.load(pkl_file, encoding='latin1'). This should solve the issue.
- When you encounter an error using old Pickle file generated using Python 2:
- Update: added a function to plot 100 MCMC examples within the 1-sigma range for posteriors (See the last figure).
- In
emcee_radex.py, change the numbers between line 404-406. - In
emcee_radex_2comp.py, change the numbers between line 540-543.
To use myradex (which may be more stable as mentioned in Ginsberg et al. 2016) instead of RADEX, please uncomment the following line:
import pyradex.fjdu
You may also need to change pyradex.Radex to pyradex.fjdu.Fjdu , and remove validate_colliders=False.
README.md: this file;emceeradex_moldata: the molecular data;results: the pickle files storing the MCMC results;single: results of one-component fittings;double: results of two-component fittings;
emcee_radex.py: one-component fitting code;emcee_radex_2comp.py: warm + cold components, in prior Tcold < Twarm; SizeCold>SizeWarm;
dataflux.note: explaining the data in Yang+2017;flux.dat: this is the flux data file read byemcee_radex.py;flux_for2p.dat: flux data used inemcee_radex_2comp.py;
A full example of a two-component fitting, the SLED plot and the corner plots:
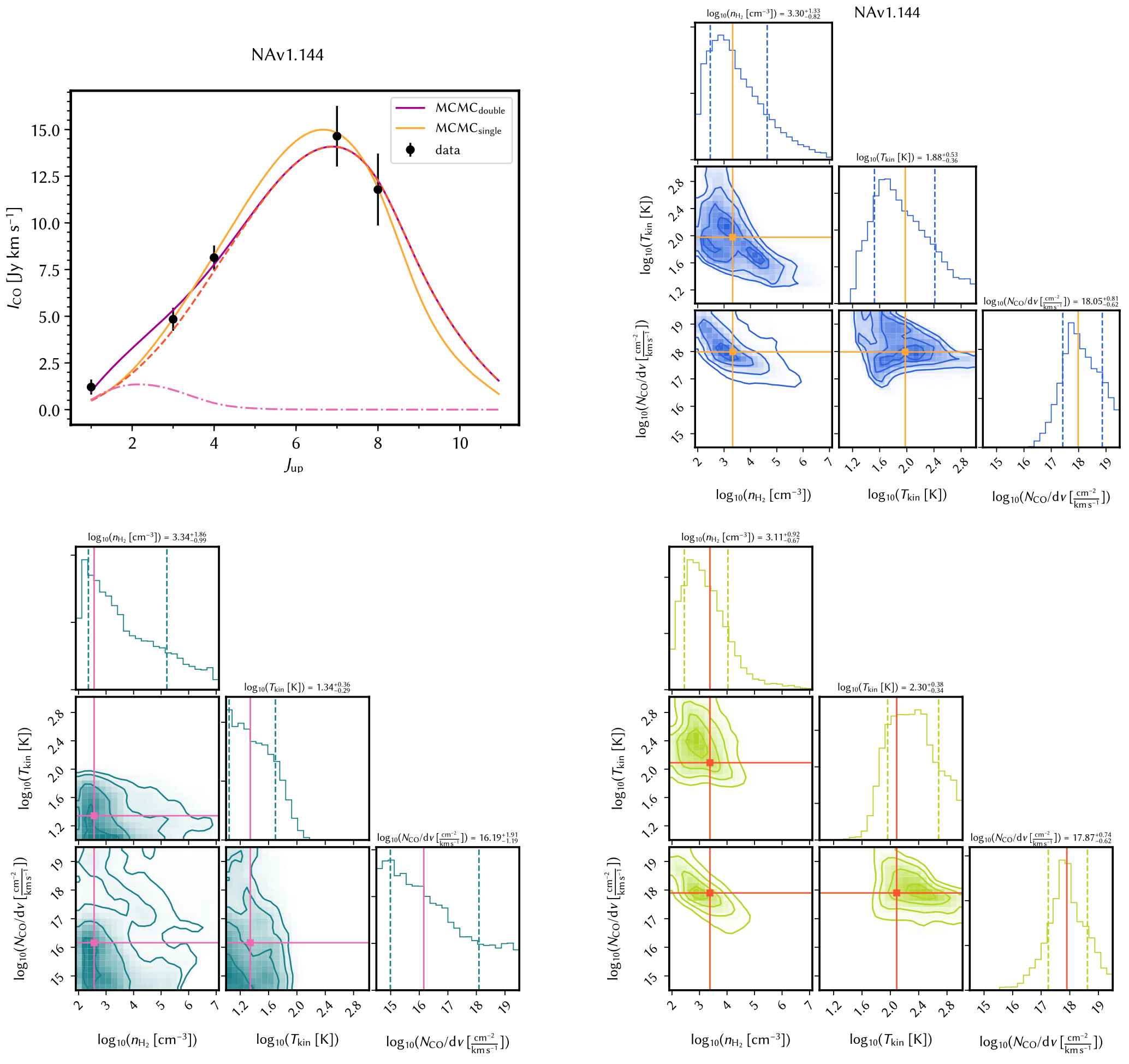
Here gives an example of the updated plot style (see notes on How to produce resulting plots):

Please cite our paper if you find this code useful. The paper also includes CO SLED data for 16 high-redshift submillimeter galaxies: C. Yang, A. Omont, A. Beelen et al. 2017, A&A, 608, A144.
Bibtex:
@ARTICLE{2017A&A...608A.144Y,
author = {{Yang}, C. and {Omont}, A. and {Beelen}, A. and {Gao}, Y. and
{van der Werf}, P. and {Gavazzi}, R. and {Zhang}, Z. -Y. and
{Ivison}, R. and {Lehnert}, M. and {Liu}, D. and {Oteo}, I. and
{Gonz{\'a}lez-Alfonso}, E. and {Dannerbauer}, H. and {Cox}, P. and
{Krips}, M. and {Neri}, R. and {Riechers}, D. and {Baker}, A.~J. and
{Micha{\l}owski}, M.~J. and {Cooray}, A. and {Smail}, I.},
title = "{Molecular gas in the Herschel-selected strongly lensed submillimeter galaxies at z 2-4 as probed by multi-J CO lines}",
journal = {\aap},
keywords = {galaxies: high-redshift, galaxies: ISM, infrared: galaxies, submillimeter: galaxies, radio lines: ISM, ISM: molecules, Astrophysics - Astrophysics of Galaxies},
year = 2017,
month = dec,
volume = {608},
eid = {A144},
pages = {A144},
doi = {10.1051/0004-6361/201731391},
archivePrefix = {arXiv},
eprint = {1709.04740},
primaryClass = {astro-ph.GA},
adsurl = {https://ui.adsabs.harvard.edu/abs/2017A&A...608A.144Y},
adsnote = {Provided by the SAO/NASA Astrophysics Data System}
}
Alexandre Beelen (ORCID: 0000-0003-3201-0185); Chentao Yang (ORCID: 0000-0002-8117-9991).