SSIM-Edge-CAN-U2Net:Two Stage Residual Learning of Reconstructing Natural Scene Images Through Multimode Fiber
This repo contains the official implementation for the paper SSIM-Edge-CAN-U2Net:Two Stage Residual Learning of Reconstructing Natural Scene Images Through Multimode Fiber.
by Le Yang (2019212184@bupt.edu.cn).
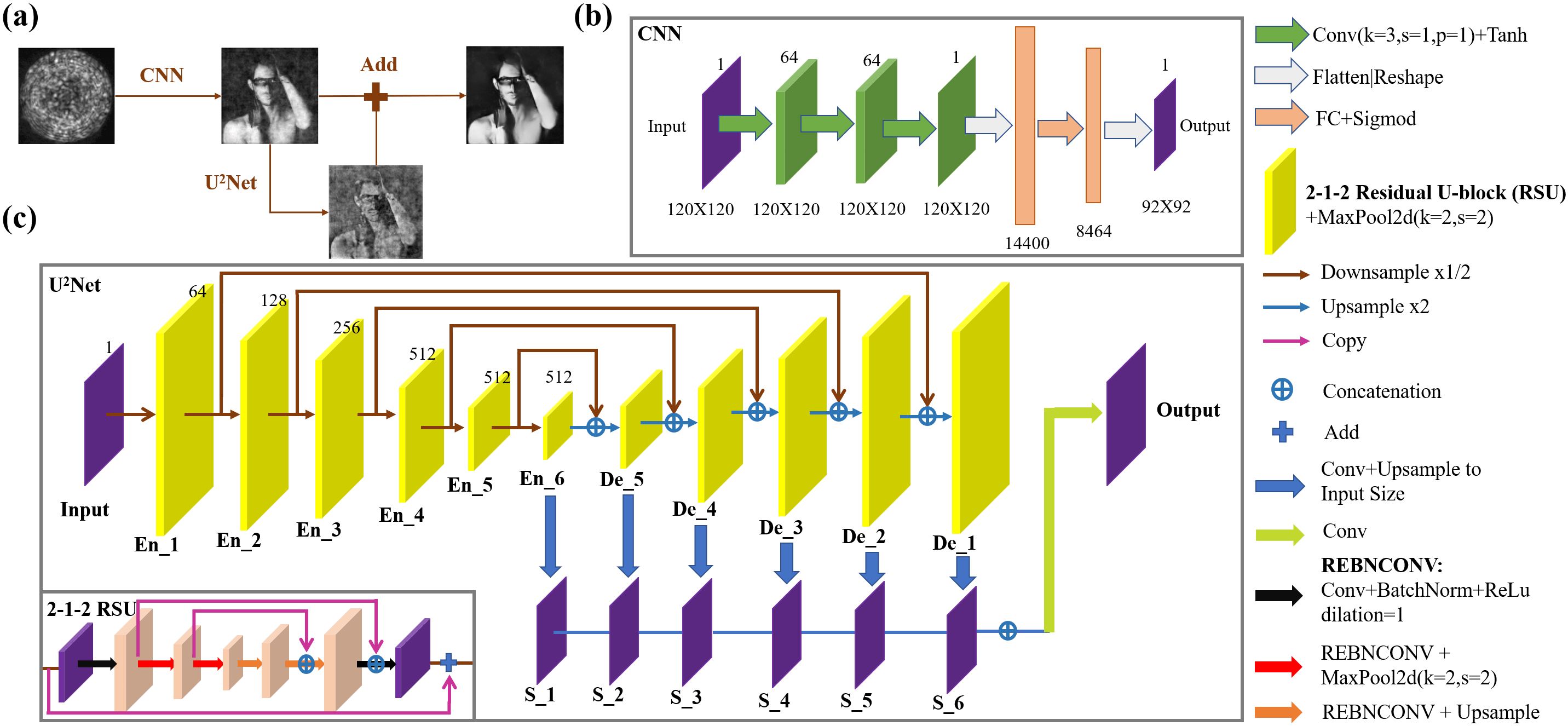
The purpose of this article is to reconstruct the natural scene images from speckles generated by multimode fiber (MMF). We use 50, 000 speckles generated by ImageNet dataset through MMF as experimental dataset. We firstly compare the different effects of MSE, SSIM and SSIM-Edge loss functions. Experiment shows that SSIM-Edge loss function performs better in many aspects. More experiments have shown that images restored by a fully convolutional network (FCN) will be blurry, while images restored by a convolutional network end with full connected layer (CAN) will be noisy. In order to make MMF speckle imaging more clearer, we propose the SSIM-Edge-CAN-U2Net model which is a two stages residual learning method. The experimental results show that the average value of the train dataset using SSIM as the evaluation index of the restored images reach 0.8359, which is 61.96% higher than that CAN model using MSE as the loss function of 0.5161, and 0.6827 (0.5154) on the test dataset, which is 32.46% higher
We use the code experiment environment conditions as shown in the following list:
- PyTorch 1.11.0
- Python 3.8 (ubuntu20.04)
- Cuda 11.3
- RTX 3090 (24GB) * 1
- Jupyter Notebook or Lab
You can install whatever python packages you find missing when running code, or just run the following command in the terminal.
pip install -r requirements.txt
pip install -r requirements.locked.txttrain-com.ipynb is the file for training the different model, test-com.ipynb is the file for testing the trained model which has been saved as *.pkl.
├── __init__.py
├── data # download from google drive
│ └── Data_1m.h5 # mmf speckle and images
├── datasets # encapsulation class for Datasets
│ ├── __init__.py
│ ├── identity.py
│ ├── normalize_0_1.py
│ ├── normalize__1_1.py
├── loss
│ ├── __init__.py # class for ssim psnr edge loss
├── model # pretrained model download from google drive
│ ├── cann-model.pkl
│ ├── epoch-ssim-175.pkl
│ ├── ssim-cann-model.pkl
│ └── u2net-ann-model.pkl
├── nets # encapsulation class for CNN 、Unet、U2net and Swin-Transformer Networks
│ ├── __init__.py
│ ├── ann.py
│ ├── can.py
│ ├── cnn_u2net.py
│ ├── swin_transformer.py
│ ├── swin_transformer_unet.py
│ ├── u2net_ann.py
│ └── unet_ann.py
├── trains # method of unified training model
│ ├── __init__.py
├── tests # method of test trained model
│ ├── __init__.py
│ ├── can.py
│ ├── cnn_u2net.py
│ └── u2net_ann.py
├── utils # method of display and evaluate results
│ ├── __init__.py
├── test-com.ipynb # the entry point of test
├── train-com.ipynb # the entry point of training
├── assets # store results
│ └── t.jpg # reconstruct images of train dataset
│ └── tt.jpg # reconstruct images of test dataset
│ └── to.jpg # reconstruct images of validation datasetAs for the training method of the model in another service and code, we will gradually move it to that repo.
Original data for 1 m MMF speckle at https://doi.org/10.5525/gla.researchdata.751. Download and place into data folder.
Link: https://drive.google.com/drive/folders/14aNid5iSMKFj9R_T-CY5-1omyewJ9dL2?usp=sharing
These checkpoint files should be placed in the correct folder in Project Structure. Download and place into model folder.
This repo partially use U2net model in U2Net repo and Unet mode in CycleGAN-and-pix2pix repo.
If you find the code/idea useful for your research, please consider citing