Installing on Linux is recommended (we have used Ubuntu 16.04).
An nVIDIA graphics card with >10GB of ram (we have used an nVIDIA Titan X Pascal) with current drivers installed (we have used nVIDIA driver version 390.48).
- Install Miniconda if necessary.
- Create a Conda environment for the platform:
conda env create -f environment.yml- Activate the environment:
conda activate fnet- Try executing the test script:
./scripts/test_run.shThe installation was successful if the script executes without errors.
Data is available as compressed tar achives here. Download and untar an image archive to the ./data/ directory (for example ./data/beta_actin/ should be full of images). All data can be automatically downloaded and untarred to the correct location by running
./scripts/paper/download_all_data.shImportant note: To build the DNA model, all data must be downloaded, as the we train on the DNA channels across all of these images.
Activate the environment if necessary (conda activate fnet). Start training a model with:
./scripts/train_model.sh dna 0The first time this is run, the DNA dataset will be split into 25% test and 75% training images. A model will be trained using the training images. This should take ~16 hours but may vary significantly depending on your system. The model will be stored in directory saved_models/dna, and there should be a run.log file whose last entries should look similar to this:
$ tail run.log
2018-02-06 16:40:24,520 - model saved to: saved_models/dna/model.p
2018-02-06 16:40:24,520 - elapsed time: 56481.3 s
2018-02-06 16:49:59,591 - BufferedPatchDataset buffer history: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29]
2018-02-06 16:49:59,592 - loss log saved to: saved_models/dna/losses.csv
2018-02-06 16:49:59,592 - model saved to: saved_models/dna/model.p
2018-02-06 16:49:59,592 - elapsed time: 57056.4 s
2018-02-06 16:59:31,301 - BufferedPatchDataset buffer history: [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29]
2018-02-06 16:59:31,302 - loss log saved to: saved_models/dna/losses.csv
2018-02-06 16:59:31,302 - model saved to: saved_models/dna/model.p
2018-02-06 16:59:31,303 - elapsed time: 57628.1 sand a losses.csv file whose last entries should look similar to this:
$ tail losses.csv
49991,0.25850439071655273
49992,0.2647261321544647
49993,0.283742755651474
49994,0.311653733253479
49995,0.30210474133491516
49996,0.2369609922170639
49997,0.2907244861125946
49998,0.23749516904354095
49999,0.3207407295703888
50000,0.3556152284145355You can train other models by replacing dna with the names of the other structures datasets (e.g., alpha_tubulin, dic_lamin_b1, fibrillarin, etc.).
./scripts/predict.sh dna 0
Predicted outputs will be in directories results/dna/test and results/dna/train corresponding to predictions on the training set and on the test set respectively. Each output directory will have files similar to this:
$ ls results/3d/dna/test
00 01 02 03 04 05 06 07 08 09 10 11 12 13 14 15 16 17 18 19 predict_options.json predictions.csvEach number above is a directory corresponding to a single dataset item (an image pair) and should have contents similar to:
$ ls results/3d/dna/test/00
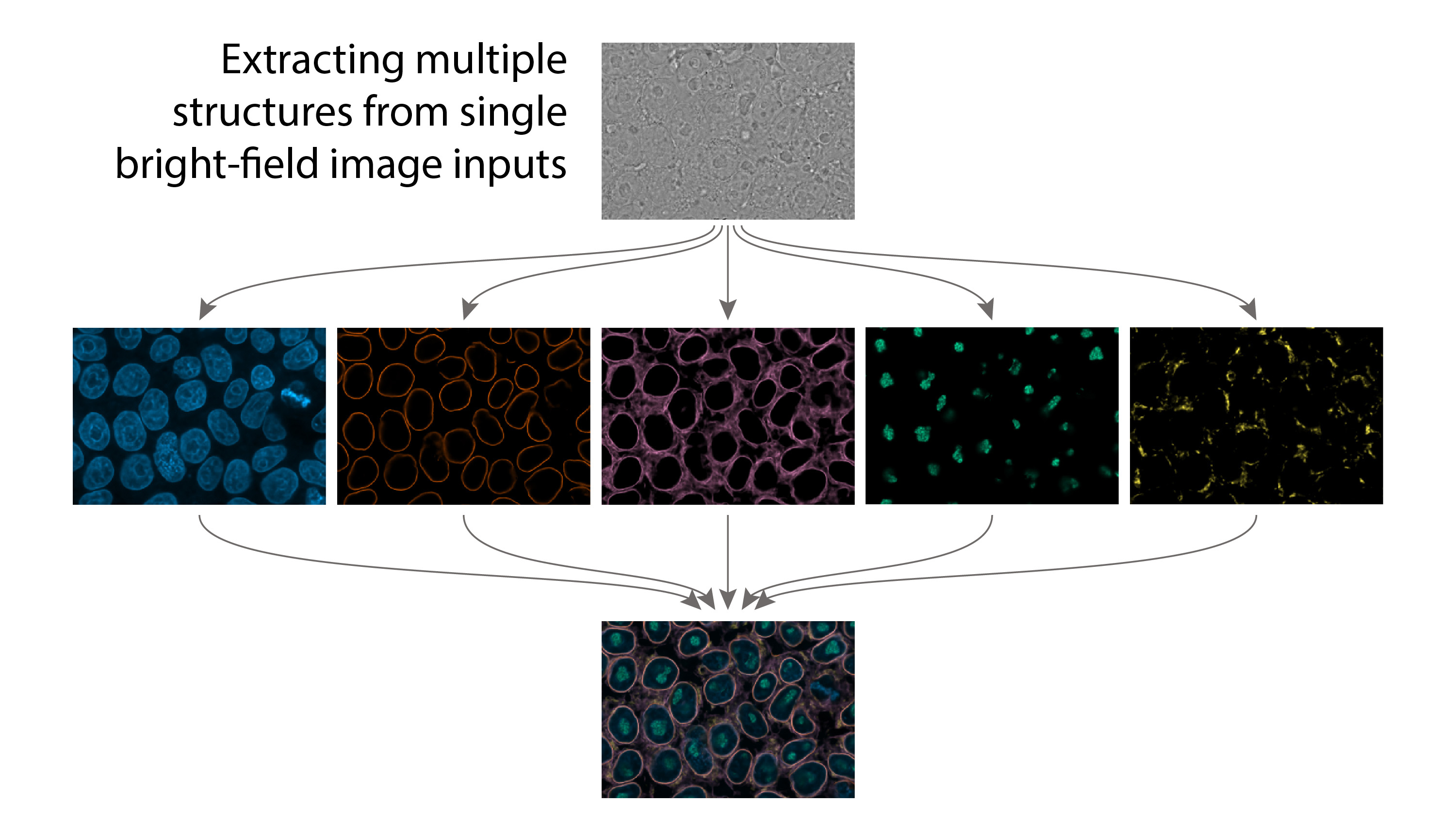
prediction_dna.tiff signal.tiff target.tiffsignal.tiff, target.tiff, and prediction_dna.tiff correspond to the input image (bright-field), the target image (real fluorescence image), and the model's output (predicted, "fake" fluorescence image) respectively.
The most general solution is to implement a new PyTorch dataset object that is responsible for loading signal images (transmitted light) and target images (fluorescence) into a consistent format. See fnet/data/tiffdataset.py or fnet/data/czidataset.py as examples. Our existing wrapper scripts will work if you make this dataset object have an __init__ function that can be correctly called with a simple keyword argument of path_csv, which points to a CSV file (example: data/csvs/mydata.csv) that describes your dataset. You should implement __getitem__() to return a PyTorch Tensor objects, where the first element is the signal data and the second element is the target image. The Tensors should be of dimensions of 1,Z,Y,X. Place your new dataset object (example: mydataset.py) in fnet/data/.
If you have single channel tiff stacks for both input and target images, you can simply use our existing tiffdataset class with a CSV that has columns labeled path_target and path_signal and whose elements are paths to where those images.
Create a new training wrapper script that is a modification of scripts/train_model.sh. Let's call it scripts/train_mymodel.sh:
#!/bin/bash -x
DATASET=${1:-dna}
N_ITER=50000
BUFFER_SIZE=30
BATCH_SIZE=24
RUN_DIR="saved_models/${DATASET}"
PATH_DATASET_ALL_CSV="data/csvs/${DATASET}.csv"
PATH_DATASET_TRAIN_CSV="data/csvs/${DATASET}/train.csv"
GPU_IDS=${2:-0}
python scripts/python/split_dataset.py ${PATH_DATASET_ALL_CSV} "data/csvs" --train_size 0.75 -v
python train_model.py \
--n_iter ${N_ITER} \
--class_dataset MyDataSet \
--path_dataset_csv ${PATH_DATASET_TRAIN_CSV} \
--buffer_size ${BUFFER_SIZE} \
--buffer_switch_frequency -1 \
--batch_size ${BATCH_SIZE} \
--path_run_dir ${RUN_DIR} \
--gpu_ids ${GPU_IDS}Now to train your model on your dataset you would run (assuming you only have 1 GPU on slot 0)
./scripts/train_mymodel.sh mydata 0This should save a trained model in saved_models/mydata, using a 75/25 train/test split on your data and saving the CSVs as data/csvs/mydata/test.csv and data/csvs/mydata/train.csv to reflect that split.
You should modify scripts/predict.sh to reflect your new dataset object as well by adding the
--class_dataset MyDataSet option. Save the modification as, say, scripts/predict_mymodel.sh.
You can then run predictions on your dataset by running
./scripts/predict_mymodel.sh mydata 0
which will output predictions into results/3d/mydata/test and results/3d/mydata/train (or into whatever output directory was specified in the predict_mymodel.sh script).
If you find this code useful in your research, please consider citing our pre-publication manuscript:
@article {Ounkomol289504,
author = {Ounkomol, Chawin and Seshamani, Sharmishtaa and Maleckar, Mary M and Collman, Forrest and Johnson, Gregory},
title = {Label-free prediction of three-dimensional fluorescence images from transmitted light microscopy},
year = {2018},
doi = {10.1101/289504},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2018/03/28/289504},
eprint = {https://www.biorxiv.org/content/early/2018/03/28/289504.full.pdf},
journal = {bioRxiv}
Gregory Johnson
E-mail: gregj@alleninstitute.org
This software license is the 2-clause BSD license plus clause a third clause that prohibits redistribution for commercial purposes without further permission.
Copyright © 2018. Allen Institute. All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
- Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
- Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
- Redistributions for commercial purposes are not permitted without the Allen Institute’s written permission. For purposes of this license, commercial purposes is the incorporation of the Allen Institute's software into anything for which you will charge fees or other compensation. Contact terms@alleninstitute.org for commercial licensing opportunities.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.