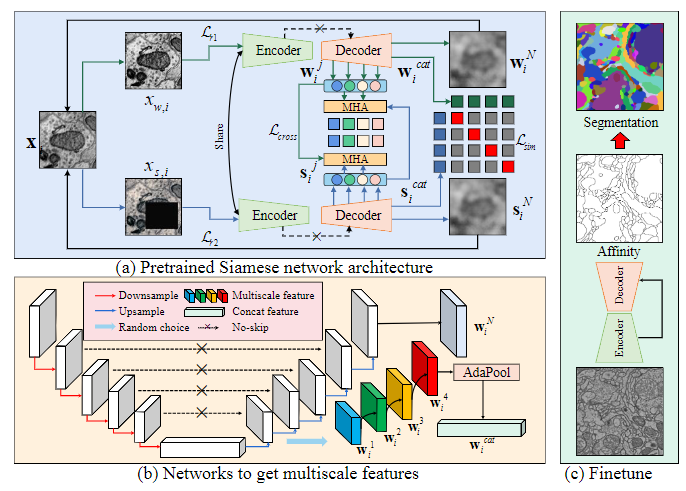
This is an official implement for Learning Multiscale Consistency for Self-supervised Electron Microscopy Instance Segmentation (ICASSP 24) Paper
To set up the required environment, you can choose from the following options:
-
Using pip: You can install the necessary Python dependencies from the
requirements.txtfile using the following command:pip install -r requirements.txt
We highly recommend using Docker to set up the required environment. Two Docker images are available for your convenience:
-
Using Docker from ali cloud:
docker pull registry.cn-hangzhou.aliyuncs.com/cyd_dl/monai-vit:v26
-
Using docker from dockerhub:
docker pull ydchen0806/cyd_docker:v1
The datasets required for pre-training and segmentation are as follows:
| Dataset Type | Dataset Name | Description | URL |
|---|---|---|---|
| Pre-training Dataset | Region of FAFB Dataset | Fly brain dataset for pre-training | EM Pretrain Dataset |
| Segmentation Dataset | CREMI Dataset | Challenge on circuit reconstruction datasets | CREMI Dataset |
| Segmentation Dataset | AC3/AC4 | AC3/AC4 Dataset | Mouse Brain GoogleDrive |
python pretrain.py -c pretraining_all -m train
python finetune.py -c seg_3d -m train -w [your pretrained path]
If you find this code or dataset useful in your research, please consider citing our paper:
@inproceedings{chen2024learning,
title={Learning multiscale consistency for self-supervised electron microscopy instance segmentation},
author={Chen, Yinda and Huang, Wei and Liu, Xiaoyu and Deng, Shiyu and Chen, Qi and Xiong, Zhiwei},
booktitle={ICASSP 2024-2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP)},
pages={1566--1570},
year={2024},
organization={IEEE}
}
If you need any help or are looking for cooperation feel free to contact us. cyd0806@mail.ustc.edu.cn