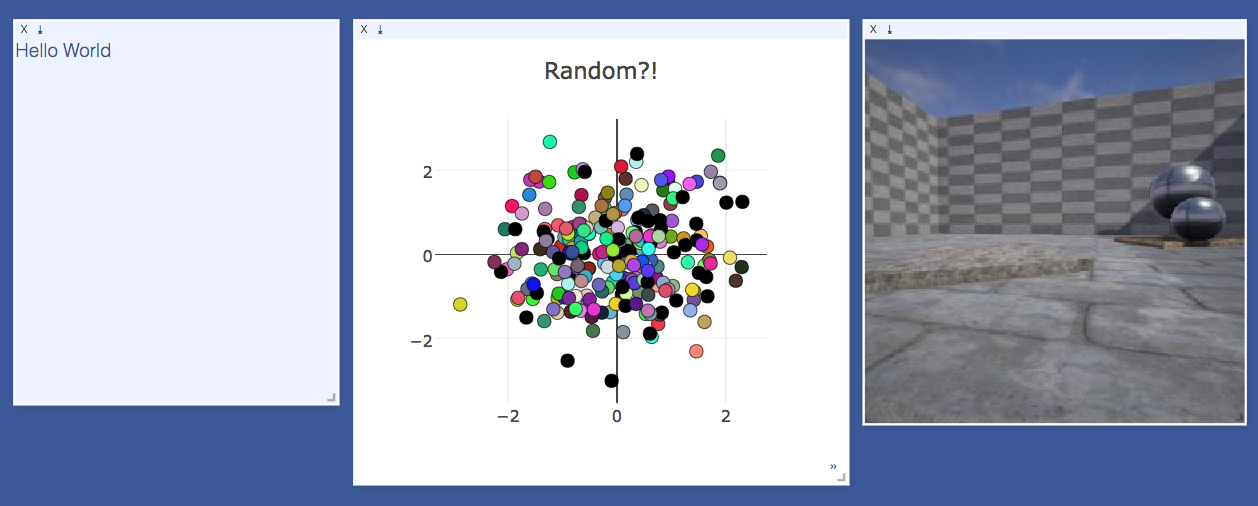
A flexible tool for creating, organizing, and sharing visualizations of live, rich data. Supports Torch and Numpy.
Visdom aims to facilitate visualization of (remote) data with an emphasis on supporting scientific experimentation.
Broadcast visualizations of plots, images, and text for yourself and your collaborators.
Organize your visualization space programmatically or through the UI to create dashboards for live data, inspect results of experiments, or debug experimental code.
Visdom has a simple set of features that can be composed for various use-cases.
The UI begins as a blank slate -- you can populate it with plots, images, and text. These appear in panes that you can drag, drop, resize, and destroy. The panes live in envs and the state of envs is stored across sessions. You can download the content of panes -- including your plots in svg.
Tip: You can use the zoom of your browser to adjust the scale of the UI.
You can partition your visualization space with envs. By default, every user will have an env called main. New envs can be created in the UI or programmatically. The state of envs is chronically saved.
You can access a specific env via url: http://localhost.com:8097/env/main. If your server is hosted, you can share this url so others can see your visualizations too.
Managing Envs: Your envs are loaded at initialization of the server, by default from
$HOME/.visdom/. Custom paths can be passed as a cmd-line argument. Envs are removed by deleting the corresponding.jsonfile from the env dir.
Once you've created a few visualizations, state is maintained. The server automatically caches your visualizations -- if you reload the page, your visualizations reappear.
-
Save: You can manually do so with the
savebutton. This will serialize the env's state (to disk, in JSON), including window positions. You can save anenvprogrammatically.
This is helpful for more sophisticated visualizations in which configuration is meaningful, e.g. a data-rich demo, a model training dashboard, or systematic experimentation. This also makes them easy to share and reuse. -
Fork: If you enter a new env name, saving will create a new env -- effectively forking the previous env.
Requires Python 2.7/3 (and optionally Torch7)
# Install Python server and client
pip install visdom
# Install Torch client
luarocks install visdom
Start the server (probably in a screen or tmux) :
python -m visdom.serverVisdom now can be accessed by going to http://localhost:8097 in your browser, or your own host address if specified.
If the above does not work, try using a SSH tunnel to your server by adding the following line to your local
~/.ssh/config:LocalForward 127.0.0.1:8097 127.0.0.1:8097.
import visdom
import numpy as np
vis = visdom.Visdom()
vis.text('Hello, world!')
vis.image(np.ones((10, 10, 3)))require 'image'
vis = require 'visdom'()
vis:text{text = 'Hello, world!'}
vis:image{img = image.fabio()}python example/demo.py
th example/demo1.lua
th example/demo2.luaThe following API is currently supported. Visualizations are powered by Plotly.
vis.scatter: 2D or 3D scatter plotsvis.line: line plotsvis.stem: stem plotsvis.heatmap: heatmap plotsvis.bar: bar graphsvis.hist: histogramsvis.boxplot: boxplotsvis.surf: surface plotsvis.contour: contour plotsvis.quiver: quiver plotsvis.image: imagesvis.text: text boxvis.save: serialize state
Further details on each of these functions are given below. For a quick introduction into the capabilities of visdom, have a look at the example directory, or read the details below.
The exact inputs into the plotting functions vary, although most of them take as input a tensor X than contains the data and an (optional) tensor Y that contains optional data variables (such as labels or timestamps). All plotting functions take as input a optional win that can be used to plot into a specific window; each plotting function also returns the win of the window it plotted in. One can also specify the env to which the visualization should be added.
This function draws a 2D or 3D scatter plot. It takes as input an Nx2 or
Nx3 tensor X that specifies the locations of the N points in the
scatter plot. An optional N tensor Y containing discrete labels that
range between 1 and K can be specified as well -- the labels will be
reflected in the colors of the markers.
The following options are supported:
options.colormap: colormap (string; default ='Viridis')options.markersymbol: marker symbol (string; default ='dot')options.markersize: marker size (number; default ='10')options.markercolor: color per marker. (torch.*Tensor; default =nil)options.legend:tablecontaining legend names
options.markercolor is a Tensor with Integer values. The tensor can be of size N or N x 3 or K or K x 3.
- Tensor of size
N: Single intensity value per data point. 0 = black, 255 = red - Tensor of size
N x 3: Red, Green and Blue intensities per data point. 0,0,0 = black, 255,255,255 = white - Tensor of size
KandK x 3: Instead of having a unique color per data point, the same color is shared for all points of a particular label.
This function draws a line plot. It takes as input an N or NxM tensor
Y that specifies the values of the M lines (that connect N points)
to plot. It also takes an optional X tensor that specifies the
corresponding x-axis values; X can be an N tensor (in which case all
lines will share the same x-axis values) or have the same size as Y.
The following options are supported:
options.fillarea: fill area below line (boolean)options.colormap: colormap (string; default ='Viridis')options.markers: show markers (boolean; default =false)options.markersymbol: marker symbol (string; default ='dot')options.markersize: marker size (number; default ='10')options.legend:tablecontaining legend names
This function draws a stem plot. It takes as input an N or NxM tensor
X that specifies the values of the N points in the M time series.
An optional N or NxM tensor Y containing timestamps can be specified
as well; if Y is an N tensor then all M time series are assumed to
have the same timestamps.
The following options are supported:
options.colormap: colormap (string; default ='Viridis')options.legend:tablecontaining legend names
This function draws a heatmap. It takes as input an NxM tensor X that
specifies the value at each location in the heatmap.
The following options are supported:
options.colormap: colormap (string; default ='Viridis')options.xmin: clip minimum value (number; default =X:min())options.xmax: clip maximum value (number; default =X:max())options.columnnames:tablecontaining x-axis labelsoptions.rownames:tablecontaining y-axis labels
This function draws a regular, stacked, or grouped bar plot. It takes as
input an N or NxM tensor X that specifies the height of each of the
bars. If X contains M columns, the values corresponding to each row
are either stacked or grouped (dependending on how options.stacked is
set). In addition to X, an (optional) N tensor Y can be specified
that contains the corresponding x-axis values.
The following plot-specific options are currently supported:
options.columnnames:tablecontaining x-axis labelsoptions.stacked: stack multiple columns inXoptions.legend:tablecontaining legend labels
This function draws a histogram of the specified data. It takes as input
an N tensor X that specifies the data of which to construct the
histogram.
The following plot-specific options are currently supported:
options.numbins: number of bins (number; default = 30)
This function draws boxplots of the specified data. It takes as input
an N or an NxM tensor X that specifies the N data values of which
to construct the M boxplots.
The following plot-specific options are currently supported:
options.legend: labels for each of the columns inX
This function draws a surface plot. It takes as input an NxM tensor X
that specifies the value at each location in the surface plot.
The following options are supported:
options.colormap: colormap (string; default ='Viridis')options.xmin: clip minimum value (number; default =X:min())options.xmax: clip maximum value (number; default =X:max())
This function draws a contour plot. It takes as input an NxM tensor X
that specifies the value at each location in the contour plot.
The following options are supported:
options.colormap: colormap (string; default ='Viridis')options.xmin: clip minimum value (number; default =X:min())options.xmax: clip maximum value (number; default =X:max())
This function draws a quiver plot in which the direction and length of the
arrows is determined by the NxM tensors X and Y. Two optional NxM
tensors gridX and gridY can be provided that specify the offsets of
the arrows; by default, the arrows will be done on a regular grid.
The following options are supported:
options.normalize: length of longest arrows (number)options.arrowheads: show arrow heads (boolean; default =true)
This function draws an img. It takes as input an CxWxH tensor img
that contains the image.
The following options are supported:
options.jpgquality: JPG quality (number0-100; default = 100)
This function prints text in a box. It takes as input an text string.
No specific options are currently supported.
The plotting functions take an optional options table as input that can be used to change (generic or plot-specific) properties of the plots. All input arguments are specified in a single table; the input arguments are matches based on the keys they have in the input table.
The following options are generic in the sense that they are the same for all visualizations (except plot.image and plot.text):
options.title: figure titleoptions.width: figure widthoptions.height: figure heightoptions.showlegend: show legend (trueorfalse)options.xtype: type of x-axis ('linear'or'log')options.xlabel: label of x-axisoptions.xtick: show ticks on x-axis (boolean)options.xtickmin: first tick on x-axis (number)options.xtickmax: last tick on x-axis (number)options.xtickstep: distances between ticks on x-axis (number)options.ytype: type of y-axis ('linear'or'log')options.ylabel: label of y-axisoptions.ytick: show ticks on y-axis (boolean)options.ytickmin: first tick on y-axis (number)options.ytickmax: last tick on y-axis (number)options.ytickstep: distances between ticks on y-axis (number)options.marginleft: left margin (in pixels)options.marginright: right margin (in pixels)options.margintop: top margin (in pixels)options.marginbottom: bottom margin (in pixels)
The other options are visualization-specific, and are described in the documentation of the functions.
- Command line tool for easy systematic plotting from live logs.
- Filtering through panes with regex by title (or meta field)
- Compiling react by python server at runtime
See guidelines for contributing here.
Visdom was inspired by tools like display and relies on Plotly as a plotting front-end.