This is an example of the MutiModal MRI images Brain Tumor Segmentation
The following dependencies are needed:
- numpy >= 1.11.1
- SimpleITK >=1.0.1
- opencv-python >=3.3.0
- tensorflow-gpu ==1.8.0
- pandas >=0.20.1
- scikit-learn >= 0.17.1
1、Preprocess
- analyze the MutiModal MRI image message and Mask image label:run the dataAnaly.py function of getMaskLabelValue() and getImageSizeandSpacing().
- MutiModal Brain Tumor MRI images have fixed size (240,240,155).
- generate patch(128,128,64) tumor image and mask for Tumor Segmentation:run the data3dprepare.py.
- save patch image and mask into csv file: run the utils.py,like file trainSegmentation.csv.
- split trainSegmentation.csv into training set and test set:run subset.py.
2、Brain Tumor Segmentation
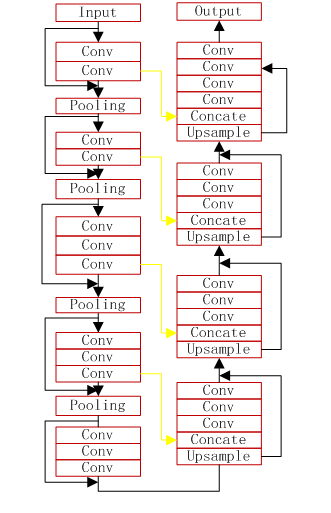
- the VNet model
- Tumor Segmentation training:run the train_Brats.py.
- Tumor Segmentation inference:run the predict_Brats.py.
- the train loss
- https://github.com/junqiangchen
- email: 1207173174@qq.com
- Contact: junqiangChen
- WeChat Number: 1207173174
- WeChat Public number: 最新医学影像技术