We computed chemical fingerprints and molecular descriptors for 2,240 compounds from OFFSIDES, 778,046 compounds from BindingDB, 8,541 compounds from Tox21. The table below shows the number of features for different fingerprint types.
| Fingerprint type | Number of features |
|---|---|
| circular | 1,024 |
| estate | 79 |
| extended | 1,024 |
| graph | 1,024 |
| hybridization | 1,024 |
| kr | 4,860 |
| maccs | 166 |
| pubchem | 881 |
| shortestpath | 1,024 |
| standard | 1,024 |
The table below shows the number of features for different descriptor types.
| Descriptor type | Number of features |
|---|---|
| constitutional | 16 |
| electronic | 35 |
| geometrical | 49 |
| hybrid | 19 |
| topological | 160 |
| all combined | 279 |
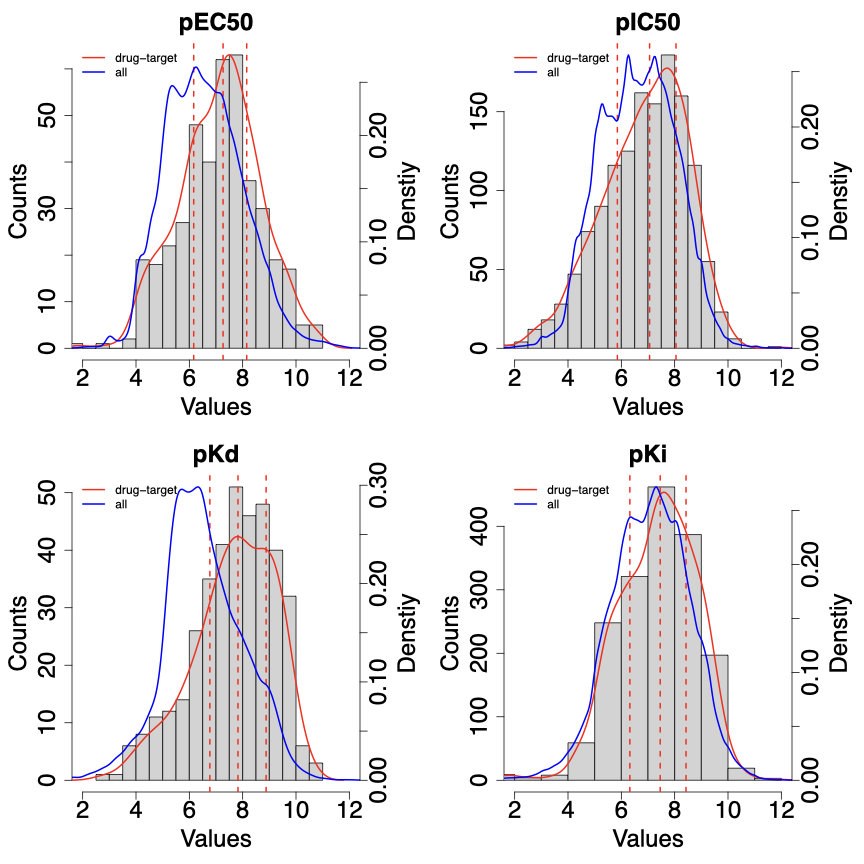
We processed compound-target relationships from BindingDB. We analyzed the distributions of binding affinity values among all known drug-target pairs from DrugBank (shown as red curves in the figure below), as compared to the distributions of binding affinity values among all compound-target pairs (shown as blue curves). We also labeled the first, second, and third quantiles of binding affinity values among all known drug-target pairs (shown as red dashed lines).
We used the first quantile as the threshold for dichotomizing binding affinity values and generating datasets of compound-target relationships. The table below shows the number of relationships for different binding affinity measurements.
| pEC50 | pIC50 | pKd | pKi | |
|---|---|---|---|---|
| positive | 44,771 | 365,419 | 7,909 | 139,521 |
| negative | 32,030 | 166,176 | 11,767 | 61,564 |
| all combined | 76,801 | 531,595 | 19,676 | 201,085 |
Detailed documentation about the source dataset can be found at downloads/. Detailed documentation about the code can be found at src/. Detailed documentation about the generated dataset can be found at data/.
-
Tatonetti NP, Patrick PY, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Science translational medicine. 2012 Mar 14;4(125):125ra31-.
-
Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, Magariños MP, Mosquera JF, Mutowo P, Nowotka M, Gordillo-Marañón M. ChEMBL: towards direct deposition of bioassay data. Nucleic acids research. 2019 Jan 8;47(D1):D930-40.
-
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic acids research. 2016 Jan 4;44(D1):D1045-53.
-
Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic acids research. 2018 Jan 4;46(D1):D1074-82.