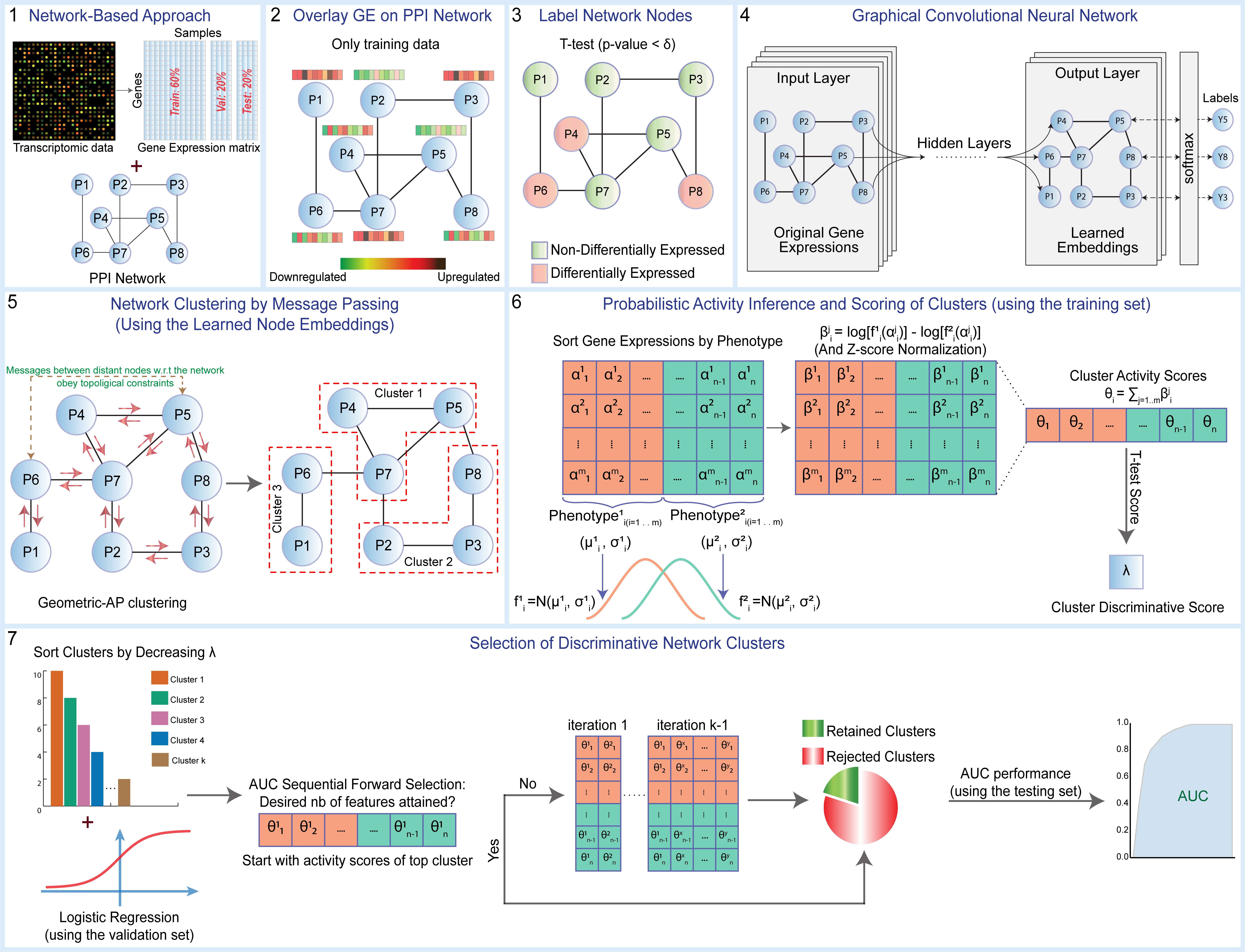
Official implementation of Graph Convolutional Network for Clustering and Classification (GCNCC).
Omar Maddouri, Xiaoning Qian, and Byung-Jun Yoon, [Deep graph representations embed network information for robust disease marker identification]
NOTE: The bash/ folder is intended to reproduce the validation and evaluation experiments from the paper.
python setup.py install
pip install -r requirements.txt
Note: The validation and evaluation pipelines under the bash/ folder contain files ordered by prefix numbers in the file names for sequential execution.
- Download the GitHub repository locally.
- Create a new folder data/ with all required sub-hierarchies as indicated in the next steps.
- Download the PPI network "9606.protein.links.v11.0.txt" or a newer version for homo sapiens from STRING (https://string-db.org/cgi/download.pl) and place it under folder data/reference/ppi_network/
- Download the dataset of interest under data/raw_input/
- For hyperparameter tuning and validation experiments, consider the pipeline under bash/validation/
- For evaluation experiments, consider the pipeline under bash/process/
Note: The output results are saved under data/output/
To be added!