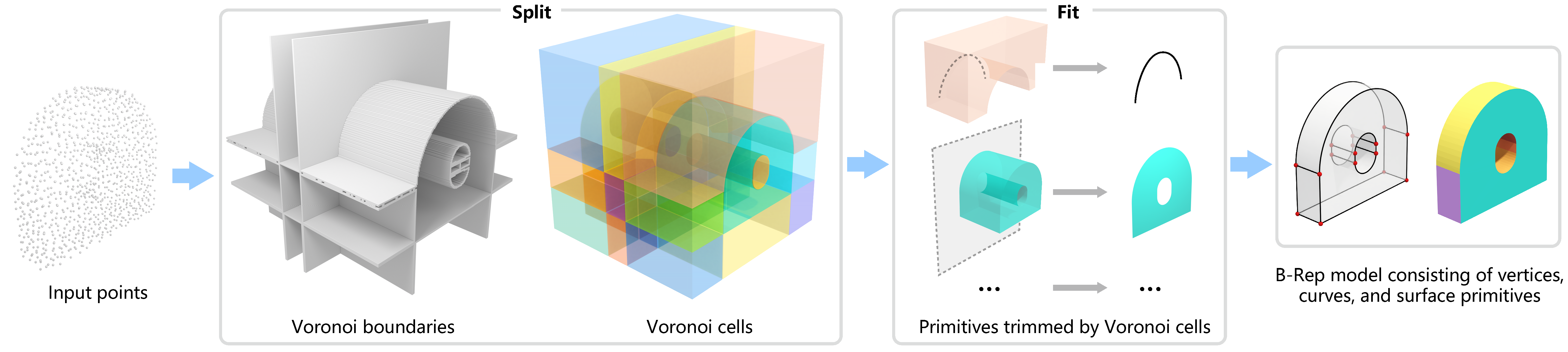
Official implementation of Split-and-Fit: Learning B-Reps via Structure-Aware Voronoi Partitioning from Yilin Liu, Jiale Chen, Shanshan Pan, Daniel Cohen-Or, Hao Zhang and Hui Huang on SIGGRAPH 2024.
NV_FF.mp4
@article {liu_sig24,
author = {Yilin Liu and Jiale Chen and Shanshan Pan and Daniel Cohen-Or and Hao Zhang and Hui Huang},
title = {{Split-and-Fit}: Learning {B-Reps} via Structure-Aware {Voronoi} Partitioning},
journal = {ACM Transactions on Graphics (Proc. of SIGGRAPH)},
volume = {43},
number = {4},
pages = {108:1--108:13},
year = {2024}
}
We provide two options for installing NVDNet: packed docker container and manual. We recommend using a packed docker container for ease of use.
Download the packed docker container including the pre-compiled environment and sample data from here (~30GB) and load it using the following command:
cat NVD.tar | docker import - nvd_release:v0
Run the container using the following command:
docker run -it --shm-size 64g --gpus all --rm --name nvdnet nvd_release:v0 /bin/zsh
If you want to connect to the container using an SSH, please refer to the docker/Dockerfile to create an SSH wrap. The default user and password is root:root.
Please refer to Dockerfile, install.sh and install_python.sh for manual installation. This script has been tested on Ubuntu 22.04 with CUDA 11.8.0.
Download the weights from here; the packed docker container has already included the weights.
Make sure you are using the latest version.
cd /root/NVDNet/build && git pull && make -j16
Put your point cloud data (w/ or w/o normal) under /data/poisson. We provide two sample point clouds in that folder.
The input folder should be organized as follows:
- root
| - test_ids.txt (the list of your filenames)
| - poisson
| | - sample.ply (your point cloud here)
Replace the /data if you use your own data
export DATA_ROOT=/data
export ROOT_PATH=/root/NVDNet
This step will generate the UDF field for the input point cloud using NDC. The output will be saved in ${DATA_ROOT}/feat/ndc.
Follow NDC's instructions if you are not using the packed container. You can speed up the process if you have multiple GPUs by replacing 1 with the number of GPUs you have. You can also use CUDA_VISIBLE_DEVICES=x to restrict the GPU usage.
cd ${ROOT_PATH} && python generate_test_voronoi_data.py ${DATA_ROOT} 1
${ROOT_PATH}/build/src/prepare_evaluate_gt_feature/prepare_evaluate_gt_feature ${DATA_ROOT}/ndc_mesh ${DATA_ROOT}/feat/ndc --resolution 256
This step will use NVDNet to predict the Voronoi diagram for the UDF field. The output Voronoi and the visualization will be saved in ${DATA_ROOT}/voronoi.
cd ${ROOT_PATH}/python && python test_model.py dataset.root=${DATA_ROOT} dataset.output_dir=${DATA_ROOT}/voronoi
This step will extract the mesh from the Voronoi diagram. The output mesh will be saved in ${DATA_ROOT}/mesh.
cd ${ROOT_PATH} && python scripts/extract_mesh_batched.py ${DATA_ROOT}/voronoi ${DATA_ROOT}/mesh ${DATA_ROOT}/test_ids.txt
Download the test data from here and extract it to /data folder in your container. Ensure the folder gt is under ${DATA_PATH}.
Run the same steps as above to generate the final mesh for evaluation. Also, specify the correct test_ids.txt when generating the mesh.
Run the evaluation script to evaluate the results.
cd ${ROOT_PATH} && ./build/src/evaluate/evaluate ${DATA_ROOT}/mesh ${DATA_ROOT}/gt /tmp/test_metrics --chamfer --matched --dist -1 --topology
We also provide the container and scripts to run HPNet, SEDNet, ComplexGen, as well as the Point2CAD. Please refer to README.md in the baselines folder.
The data preparation and training code has already been provided in the src and python folders. However, since the Voronoi diagram is a volumetric representation, the whole dataset is too large (1.5TB) to be uploaded. We provide the code to generate the Voronoi diagram from the UDF field. You can use the code to generate the Voronoi diagram for your own data.