Hi, PolyFilterPlayground is a repo for quick hyperparameter tuning, training and testing for polynomial-filter-based GNNs. It is written by Ph.D. student Yuhe Guo from Group Alg@RUC. I've summarized this repo from my own research experience, and it greatly reduces the amount of work I have to do to try out a model.
This repo is still under construction. Some features of this repo are:
- Support hyper-parameter tuning via
Optunaefficiently. Features of Optuna likePrunersare used. - The records are well-cached.
- Support most datasets (though some of them use interfaces from
PyGdirectly). - Some modules, such as
ROC-AUCis much faster than the common practice of using implementations fromscikit-learn.
Expectations. I hope this repo can help you to be productive to try out your ideas. I also hope that it can help you to check the performances of former models with your own practice (The number of models I put under the folder \model is small. It would not take much time to add more models if you want to try out. You are also welcomed to join me and contribute more models).
I write a demo to go through this repo. For more information, feel free to contact me via guoyuhe@ruc.edu.cn.
Use the following command to tune OptBasisGNN (I also call it NormalNN sometimes) on the Chameleon dataset.
To get started quickly, we only test 10 optuna trials, and 50 epochs for each trial. (In real experiments, I suggest using 100 or 200 trials, and 2000 epochs for each trial.)
python tune.py --model OptBasisGNN --dataset geom-chameleon --gpu 0 --logging --log-detailedCh --optuna-n-trials 10 --n-epochs 50Remark: Influence of this step.
-
Optuna cache. This command creates a database in
cache/OptunaTrials/. This is whereOptunagather the trial histories and upon them search for the next hyper-parameters.\cache ---\ckpts ---\OptunaTrials ------\OptBasisGNN-geom-chameleon.db -
Logging Modes. Here,
--logging --log-detailedChmeans that the detailed information of each trial will be printed in the console. You can also specify other logging forms, e.g.--logging --log-detail --id-log [folder_name], which will make it silent in the console, and instead, log the details for each trial in the folderruns/Logs[folder_name]. I put some examples incmds_tune.sh:name="gpr-cham" python tune.py --model GPRGNN --dataset geom-chameleon --gpu 0 --logging --log-detail --id-log 1011014503 1>>logs/${name}.log 2>>logs/${name}.err & sleep 1 name="opt-roman-empire" python tune.py --model NormalNN --dataset roman-empire --gpu 0 --logging --log-detail --id-log 1015014501 1>logs/${name}.log 2>logs/${name}.err &
-
How is the search sparce of hyperparameters specified? I put them in
opts/. Please read the logic ininitialize_args()intune.py. If you want to tune your own model, remember to specify some options underopts/also. (Also remember to slightly trimbuild_optimizers()andbuild_models()for your own practice.)
Following Step 1, now let the selected hyper-parameters be reported:
```bash
python report.py --model OptBasisGNN --dataset geom-chameleon --gpu 0 --logging --log-detailedCh --optuna-n-trials 10
```
The output is as follows. You can directly copy it to train!
```bash
python train.py --dropout 0.5 --dropout2 0.5 --lr1 0.05 --lr2 0.03 --n-layers 10 --wd1 1e-07 --wd2 1e-05 --seed 42 --model OptBasisGNN --dataset geom-chameleon --logging --log-detailedCh --udgraph --early-stop --n-hidden 64 --n-cv 10 --es-ckpt OptBasisGNN-geom-chameleon --log-detail --log-detailedCh 1>logs/OptBasisGNN-geom-chameleon.log 2>logs/OptBasisGNN-geom-chameleon.err&
```
Remarks:
-
The command above seem a bit verbose. Actually, most parameters like
--early-stop,--loggingand--udgraph(using undirected graph, which is a common practice in polynomial-filter based GNNs) are the same among all the practices. -
Note that if the number of
--optuna-n-trialsyou assign is larger than the finished trials in the tuning process, the result will not be reported. -
Make sure the name of
--modeland--datasetis the same in the Step 1 and Step 2. They decides the name of Optuna cache database that are you using.
Just copy the output in Step 2 to run!
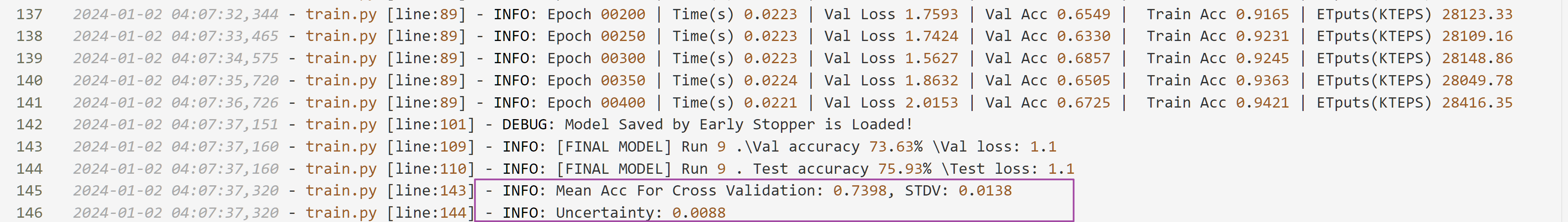
Since we specified 1>logs/OptBasisGNN-geom-chameleon.log 2>logs/OptBasisGNN-geom-chameleon.err&,
the results can be found in logs/OptBasisGNN-geom-chameleon.log.
We find that even tuned for such a small number of 10 optuna trials, and 50 epochs for each trial, the final result, 73.98±0.88 over 10 random splits is quite high.