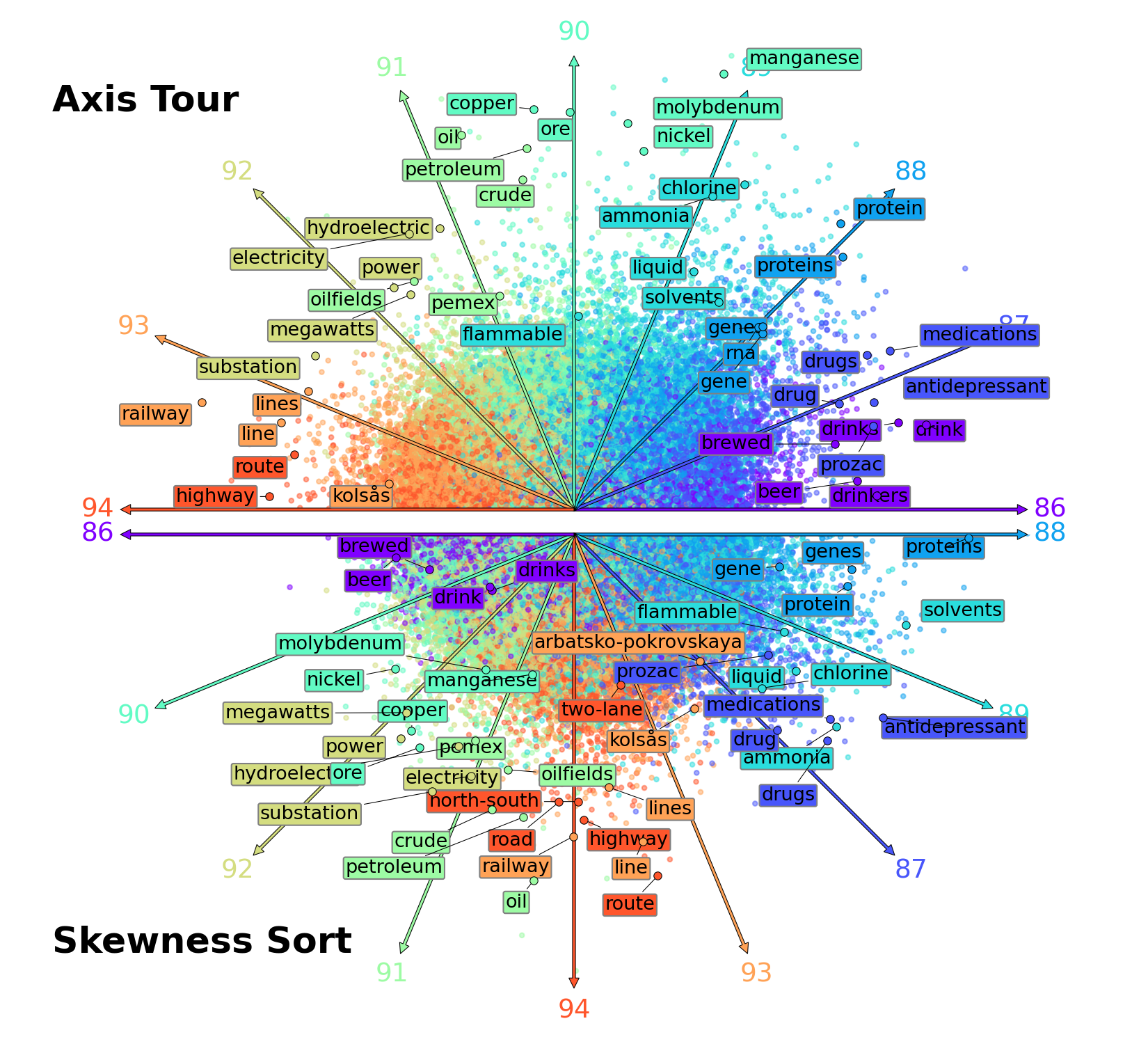
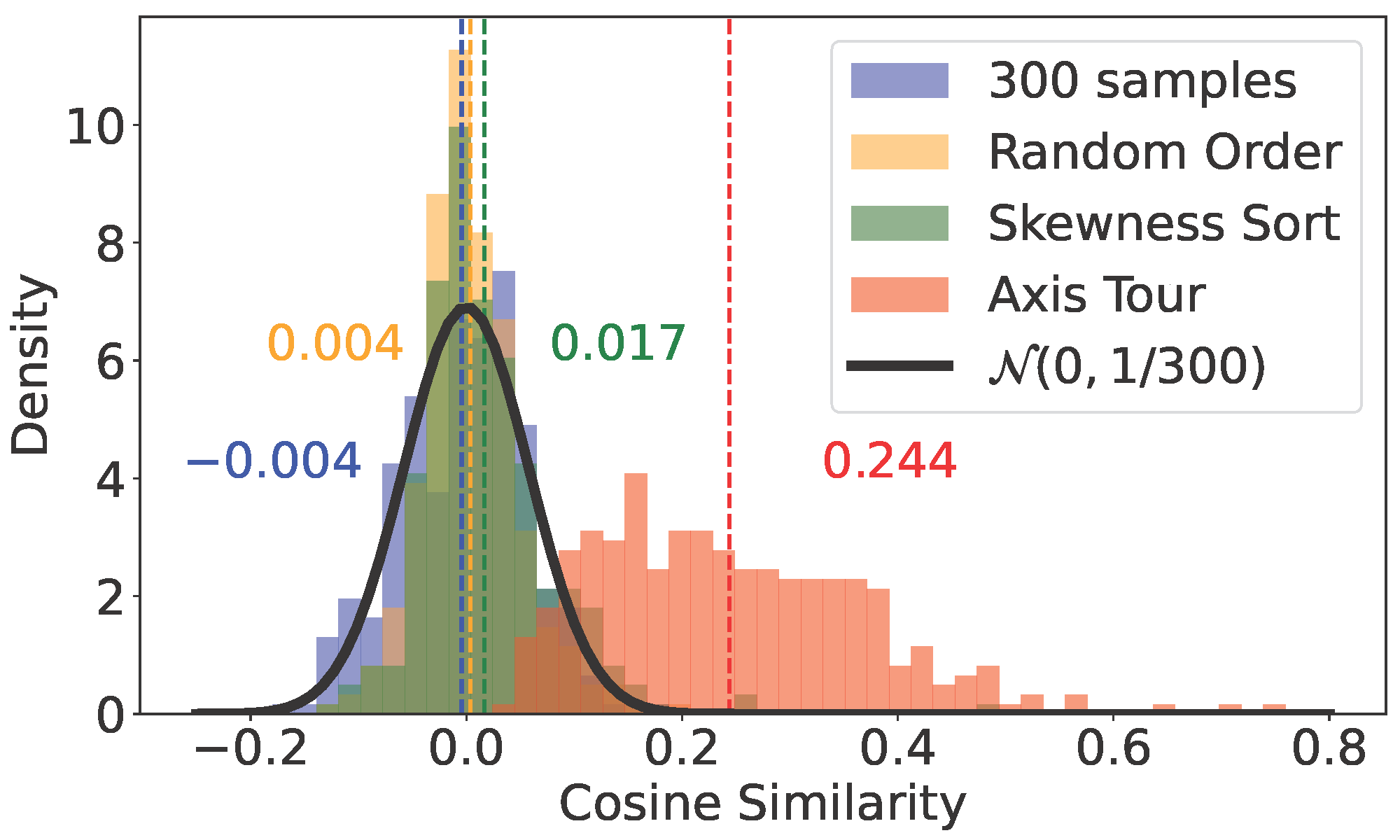
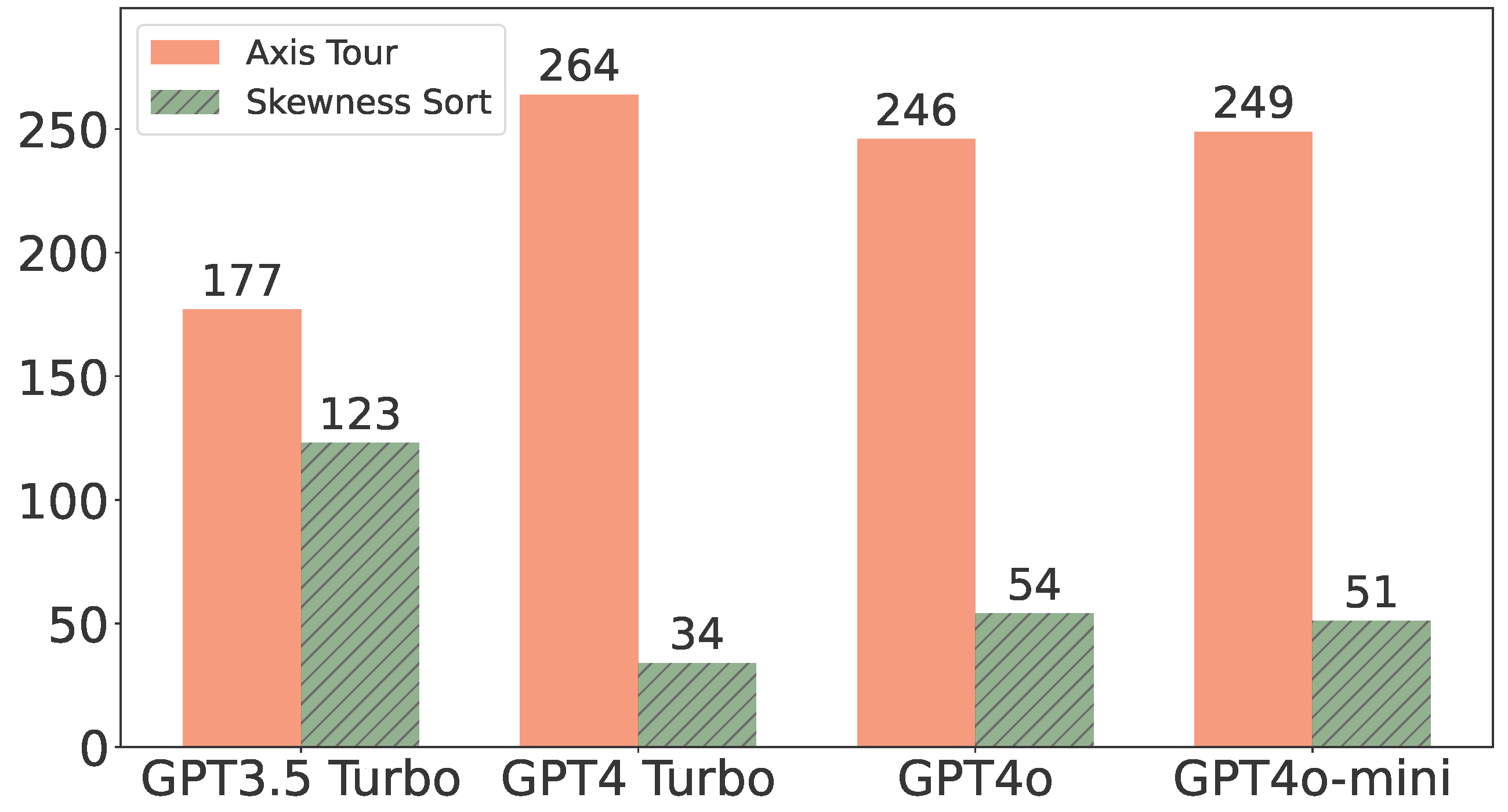
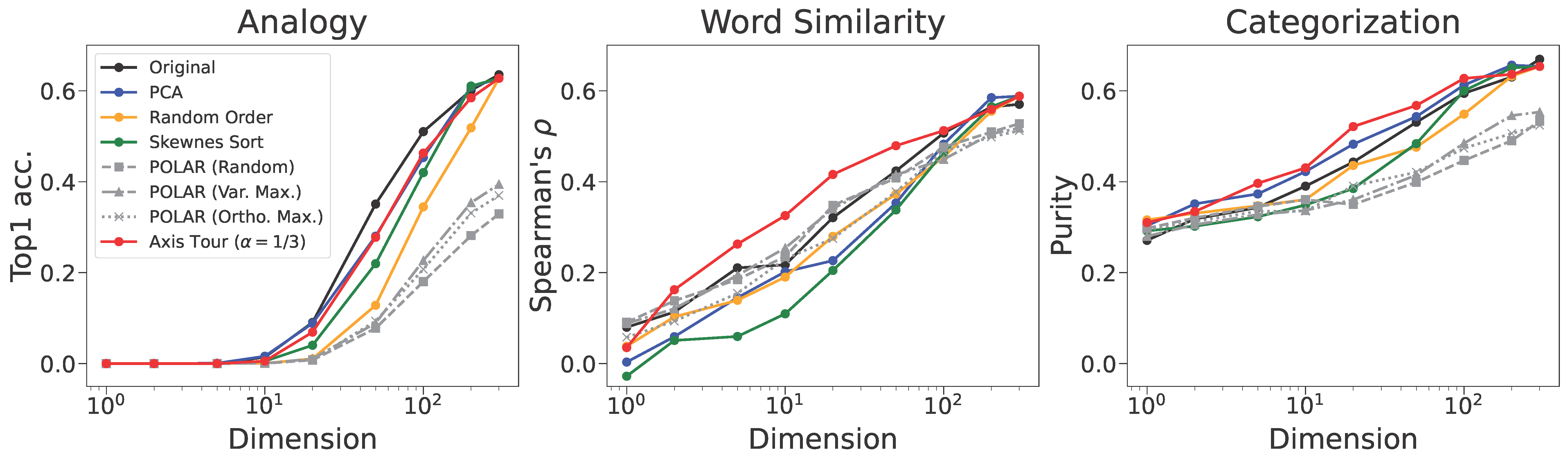
Axis Tour: Word Tour Determines the Order of Axes in ICA-transformed Embeddings
Hiroaki Yamagiwa, Yusuke Takase, Hidetoshi Shimodaira
EMNLP 2024 Findings
This repository is intended to be run in a Docker environment. If you are not familiar with Docker, please install the packages listed in requirements.txt.
Please create a Docker image as follows:
bash script/docker_build.sh# Set OPENAI API key
export OPENAI_API_KEY="sk-***"
# Set the DOCKER_HOME to specify the path of the directory to be mounted as the home directory inside the Docker container
export DOCKER_HOME="path/to/your/docker_home"bash script/docker_run.shInstead of recomputing the embeddings, you can access the embeddings used in the paper through the following links. Note that sign flip was not applied to the ICA-transformed embeddings to ensure that the skewness of the axes remains positive.
Place the downloaded file under the directory output/raw_embeddings as shown below:
$ ls output/raw_embeddings/
raw_glove.pklPlace the downloaded file under the directory output/pca_ica_embeddings/ as shown below:
$ ls output/pca_ica_embeddings/
pca_ica_glove.pkl-
GloVe (Google Drive) for
$k=100$
Place the downloaded file under the directory output/axistour_embeddings/ as shown below:
$ ls output/axistour_embeddings/
axistour_top100_glove.pklPlace the downloaded files under the directory output/polar_glove_embeddings/ as shown below:
$ ls output/polar_glove_embeddings/
orthogonal_antonymy_gl_500_StdNrml.bin rand_antonym_gl_500_StdNrml.bin variance_antonymy_gl_500_StdNrml.binCreate the data/embeddings directory:
mkdir -p data/embeddingsDownload GloVe embeddings as follows:
wget https://nlp.stanford.edu/data/glove.6B.zip
unzip glove.6B.zip -d data/embeddings/glove.6BFor more details, please refer to the original repository: stanfordnlp/GloVe.
Similar to [1], download LKH solver for TSP:
wget http://webhotel4.ruc.dk/~keld/research/LKH-3/LKH-3.0.6.tgz
tar xvfz LKH-3.0.6.tgzWe have modified the repository word-embeddings-benchmarks [5]. To install it, use the following commands:
cd word-embeddings-benchmarks
pip install -e .
cd ..Similar to [2], calculate PCA-transformed and ICA-transformed embeddings as follows:
python save_pca_and_ica_embeddings.py --emb_type glovepython save_axistour_embeddings.py --emb_type glove --topk 100This will generate the scatterplot shown above:
python make_scatterplots.py --emb_type glove --topk 100 --left_axis_index 86 --length 9If you are not using adjustText==1.0.4, you may need to manually adjust the position of the text.
To save the top 5 words of the Axis Tour embeddings (
python save_axistour_top_words.py --emb_type glove --topk 100 --top_words 5The output file will be as follows:
$ head -n 6 output/axistour_top_words/glove_top100-top5_words.csv
axis_idx,top1_word,top2_word,top3_word,top4_word,top5_word
0,phaen,sandretto,nakhchivan,burghardt,regno
1,region,goriška,languedoc,regions,saguenay-lac-saint-jean
2,mountain,mount,mountains,everest,peaks
3,stage,vinokourov,vuelta,stages,magicians
4,italy,italian,di,francesco,pietropython make_cossim_histogram.py --emb_type glove --topk 100python eval_continuity_by_OpenAI_API.pygit clone https://github.com/Sandipan99/POLAR.gitThen
cp polar_glove.ipynb POLAR/Run POLAR/polar_glove.ipynb to generate embeddings saved under output/polar_glove_embeddings/ with the filename ${method}_gl_500_StdNrml.bin.
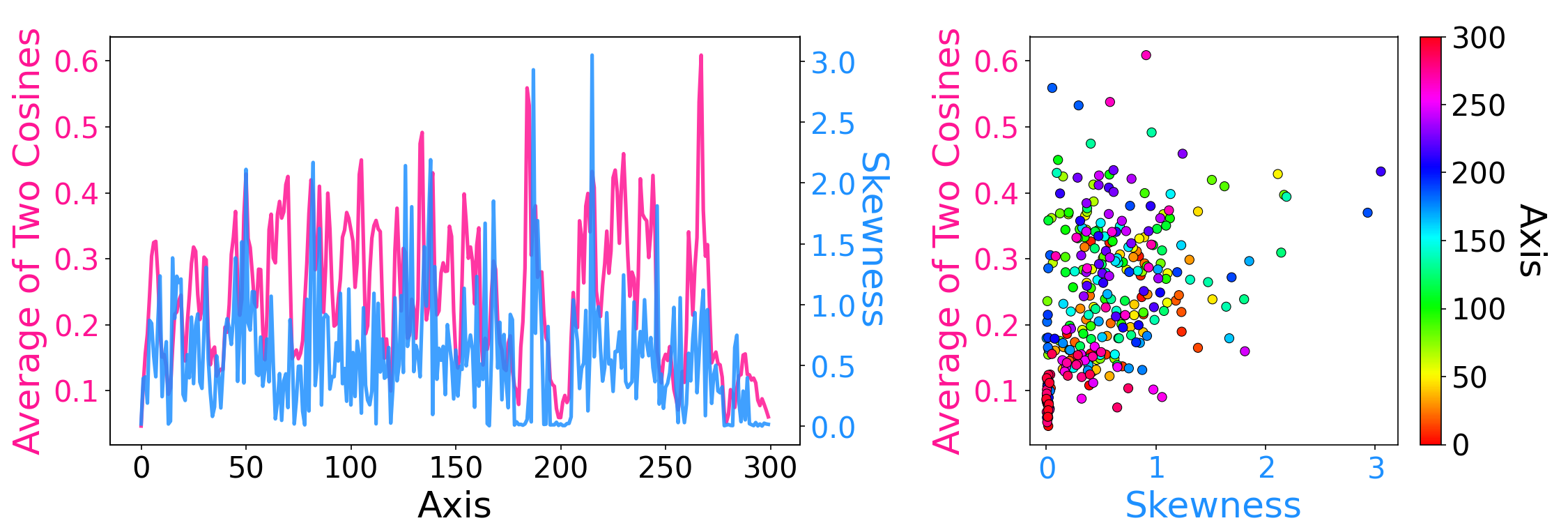
python make_dimred_figure.py --emb_type glove --fig_type main python make_relation_skewness_and_two_cossims.py --emb_type glove --topk 100| (a) Axis Tour |
|---|
| (b) Skewness Sort |
|---|
[1] Sato. Word Tour: One-dimensional Word Embeddings via the Traveling Salesman Problem. NAACL. 2022.
[2] Yamagiwa et al. Discovering Universal Geometry in Embeddings with ICA. EMNLP. 2023.
[3] Chelba et al. One billion word benchmark for measuring progress in statistical language modeling. INTER-SPEECH. 2014.
[4] Mathew, et al. The POLAR framework: Polar opposites enable interpretability of pretrained word embeddings. Web Conference. 2020.
[5] Jastrzebski et al. How to evaluate word embeddings? On importance of data efficiency and simple supervised tasks. arXiv. 2017.
If you find our code or model useful in your research, please cite our paper:
@inproceedings{yamagiwa-etal-2024-axis,
title = "Axis Tour: Word Tour Determines the Order of Axes in {ICA}-transformed Embeddings",
author = "Yamagiwa, Hiroaki and
Takase, Yusuke and
Shimodaira, Hidetoshi",
editor = "Al-Onaizan, Yaser and
Bansal, Mohit and
Chen, Yun-Nung",
booktitle = "Findings of the Association for Computational Linguistics: EMNLP 2024",
month = nov,
year = "2024",
address = "Miami, Florida, USA",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/2024.findings-emnlp.28",
pages = "477--506",
abstract = "Word embedding is one of the most important components in natural language processing, but interpreting high-dimensional embeddings remains a challenging problem. To address this problem, Independent Component Analysis (ICA) is identified as an effective solution. ICA-transformed word embeddings reveal interpretable semantic axes; however, the order of these axes are arbitrary. In this study, we focus on this property and propose a novel method, Axis Tour, which optimizes the order of the axes. Inspired by Word Tour, a one-dimensional word embedding method, we aim to improve the clarity of the word embedding space by maximizing the semantic continuity of the axes. Furthermore, we show through experiments on downstream tasks that Axis Tour yields better or comparable low-dimensional embeddings compared to both PCA and ICA.",
}
- Since the URLs of published embeddings may change, please refer to the GitHub repository URL instead of the direct URL when referencing in papers, etc.
- This directory was created by Hiroaki Yamagiwa.
- The code for TICA was created by Yusuke Takase.
- See README.Appendix.md for the experiments in the Appendix.