- pandas
- numpy
- Biopython
- tqdm
- Badread (error_simulator.py)
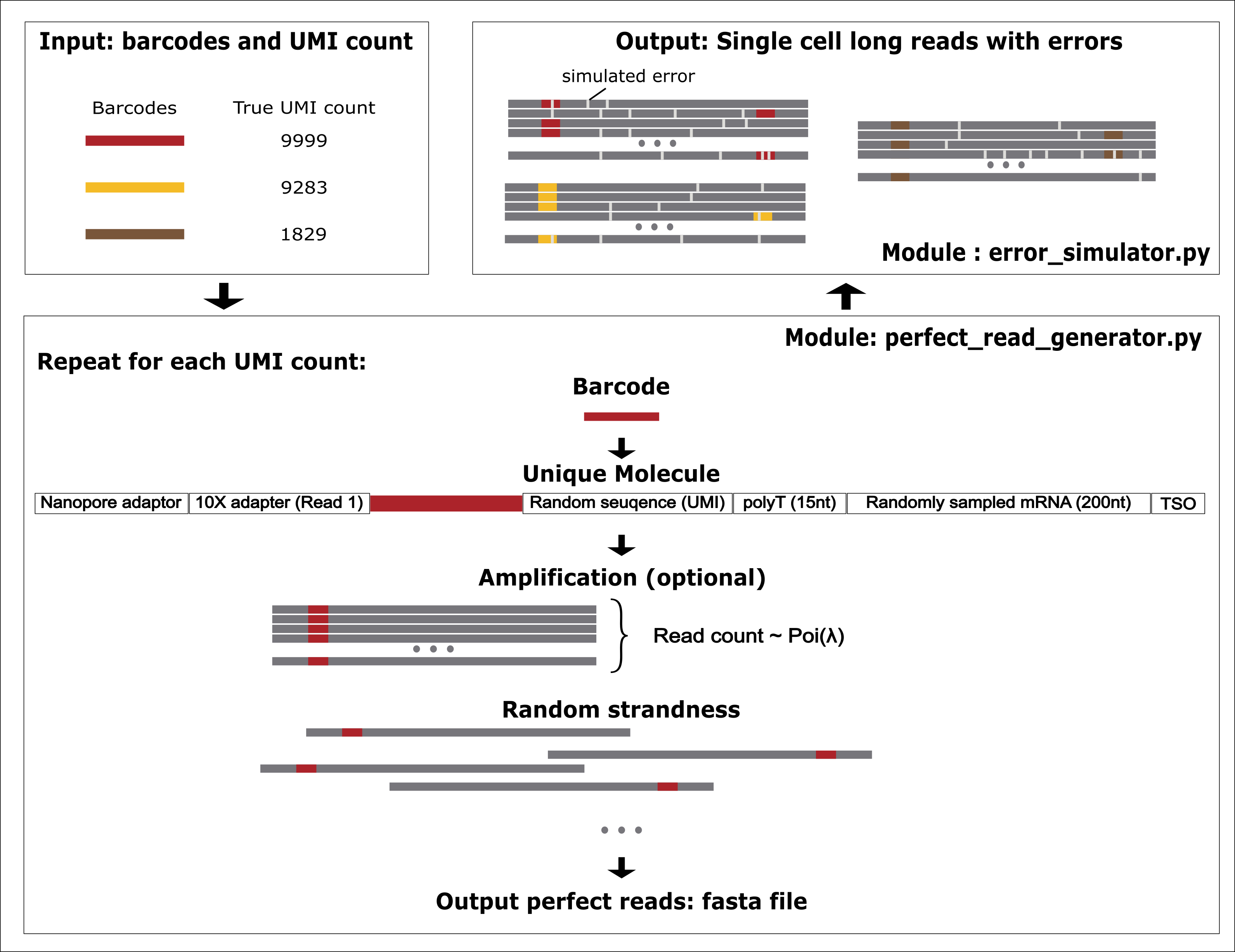
- SLSsim randomly sample cDNA from transcript reference and attach 10x cell barcodes, UMI, poly(dT) and Template switching oligo (TSO) to form perfect reads (i.e. reads with no error.):
bin/perfect_read_generator.py - SLSsim using the error model and qscore modle from Badread (one of the depencencies) to introduce error into the reads (
bin/error_simulator.py).
git clone https://github.com/youyupei/SLSim
cd SLSim
- Generate perfect reads (
template.fa)
python3 bin/perfect_read_generator.py -r reference_transcript.fa -i bc_umi.csv -o template.fa
The output from perfect_read_generator.py is a FASTA file, each entry is an error-free read with the true barcode followed by a index as read id.
- Simulate nanopore errors and create fastq file
python3 bin/error_simulator.py -t template.fa
The output from error_simulator.py is a FASTQ file, each entry is a error-containing read with per-base quality score. The read id can be matched to those in the FASTA file generated from perfect_read_generator.py.