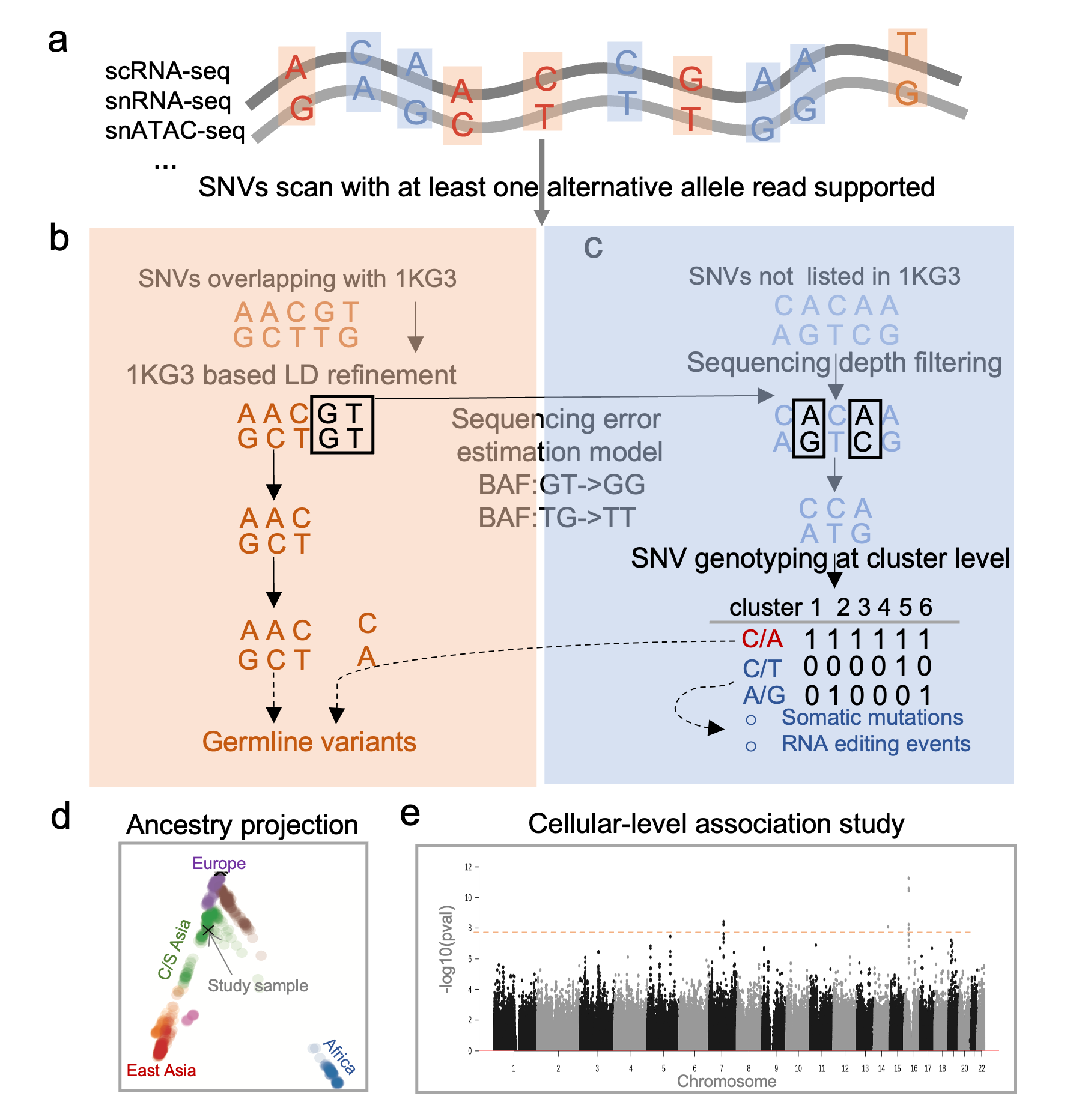
SNV calling from single cell sequencing
Monopogen is an analysis package for SNV calling from single-cell sequencing, developed and maintained by Ken chen's lab in MDACC. Monopogen is developed to benefit the population-level ancestry and association study for single cell sequencing studies. It can work on datasets generated from single cell RNA 10x 5', 10x 3', smartseq, single ATAC-seq technoloiges etc.
It is composed of four modules:
- Germline SNV identification from sparse 10x scRNA-seq or scATAC-seq profiles. Given the sparsity of single cell sequencing data, we leverage linkage disequilibrium (LD) from external reference panel(such as 1KG3, TopMed) to refine genotypes.
- Putative somatic variant or RNA editing events identification. The SNVs with high alternative allele supporeted in study sample are further classifed based on their allele frequency patten among cell clusters (cell type/ cell states). The variant calling is mostly from
Monovardeveloped in Ken chen's lab in MDACC. Theoretically, germline SNVs can be detected in most of clusters while only specfic clusters for somatic mutations.
The output of Monopogen will enable ancestry identificaiton on single cell samples and genome-wide association study on the celluar level if sample size is sufficient.
- python (version >= 3.73)
- java (open JDK>=1.8.0)
- pandas>=1.2.3
- pysam>=0.16.0.1
- NumPy>=1.19.5
- sciPy>=1.6.3
- pillow>=8.2.0
Right now Monopogen is avaiable on github, you can install it through github
git clone https://github.com/KChen-lab/Monopogen.git
cd Monopogen
pip install -e .
You can type the following command to get the help information.
python ./src/Monopogen.py SCvarCall --help
usage: Monopogen.py SCvarCall [-h] --bamFile BAMFILE --step {germline,bamFiltering,beagleImputation} --mode
{single,multiple} --chr CHR [--out OUT] --appPath APPPATH --reference REFERENCE
--imputation-panel IMPUTATION_PANEL [--depthFilter DEPTHFILTER] [--altRatio ALTRATIO]
[--wildCluster WILDCLUSTER] [--mutationCluster MUTATIONCLUSTER]
[--max-mismatch MAX_MISMATCH] [--max-softClipped MAX_SOFTCLIPPED] --cellCluster CELLCLUSTER
optional arguments:
usage: Monopogen.py SCvarCall [-h] -b BAMFILE -j STEP -y MODE -c CHR [-o OUT]
-r REFERENCE -p IMPUTATION_PANEL
[-d DEPTH_FILTER] [-l DEPTH_FILTER_MONOVAR]
[-t ALT_RATIO] [-w WILDCLUSTER]
[-u MUTATIONCLUSTER] [-m MAX_MISMATCH]
[-s MAX_SOFTCLIPPED] -a APP_PATH -i CELL_CLUSTER
optional arguments:
-h, --help show this help message and exit
-b BAMFILE, --bamFile BAMFILE
The bam file for the study sample, the bam file should
be sorted (default: None)
-j STEP, --step STEP varinat calling in germline of individual level or
somatic variants in cluster level (default: all)
-y MODE, --mode MODE single sample or multiple sample (default: None)
-c CHR, --chr CHR The chromosome used for variant calling (default:
None)
-o OUT, --out OUT The output director (default: None)
-r REFERENCE, --reference REFERENCE
The human genome reference used for alignment
(default: None)
-p IMPUTATION_PANEL, --imputation-panel IMPUTATION_PANEL
The population-level variant panel for variant
refinement such as 1000 Genome 3 (default: None)
-d DEPTH_FILTER, --depth_filter DEPTH_FILTER
The sequencing depth filter for variants not
overlapped with public database (default: 50)
-l DEPTH_FILTER_MONOVAR, --depth_filter_monovar DEPTH_FILTER_MONOVAR
The sequencing depth filter for variants not
overlapped with public database (default: 50)
-t ALT_RATIO, --alt_ratio ALT_RATIO
The minina allele frequency for variants as potential
somatic mutation (default: 0.1)
-w WILDCLUSTER, --wildCluster WILDCLUSTER
The mininal number of cluster support wilde type
(default: 2)
-u MUTATIONCLUSTER, --mutationCluster MUTATIONCLUSTER
The mininal number of cluster support mutated type
(default: 1)
-m MAX_MISMATCH, --max-mismatch MAX_MISMATCH
The maximal mismatch allowed in one reads for variant
calling (default: 3)
-s MAX_SOFTCLIPPED, --max-softClipped MAX_SOFTCLIPPED
The maximal soft-clipped allowed in one reads for
variant calling (default: 1)
-a APP_PATH, --app-path APP_PATH
The app library paths used in the tool (default: None)
-i CELL_CLUSTER, --cell_cluster CELL_CLUSTER
The cell cluster csv file used for somatic variant
calling (default: None)
We provide one demo of SNV calling based on data provided in the example/ folder, which includes:
chr20_2Mb.rh.filter.sort.bam (.bai)
The bam file storing read alignment for one study sample. CurrentMonopogensupports both single and mulitple sample calling mode. Mulitple sample calling will increase the sensitivity of variant detection.CCDG_14151_B01_GRM_WGS_2020-08-05_chr20.filtered.shapeit2-duohmm-phased.vcf.gzThe reference panel with over 3,000 samples in 1000 Genome database. Only SNVs located in chr20: 0-2Mb were extracted in this vcf file.chr20_2Mb.hg38.fa (.fai)
The genome reference used for read aligments. Only seuqences in chr20:0-20Mb were extracted in this fasta file.cell_cluster.csv
The cell cluster information. In the csv file, the first column is the cellID and the second column is the cluster (can be derived fromSeurat)
There is a bash script ./test/test.chr20.sh to run above example in the folder test:
path="XX/Monopogen"
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:${path}/apps
python ../src/Monopogen.py SCvarCall \
-b ../example/chr20_2Mb.rh.filter.sort.bam \
-j all \
-y single \
-a ../apps \
-c chr20 \
-o out \
-i ../example/cell_cluster.csv \
-p ../example/CCDG_14151_B01_GRM_WGS_2020-08-05_chr20.filtered.shapeit2-duohmm-phased.vcf.gz \
-r ../example/chr20_2Mb.hg38.fa -d 200 -t 0.1 -m 3 -s 5
The most important results are in the folder out/SCvarCall:
chr20.germline.vcf
In this vcf file, the germline SNVs were genotyped for the study sample
##fileformat=VCFv4.2
##filedate=20220808
##source="beagle.27Jul16.86a.jar (version 4.1)"
##INFO=<ID=AF,Number=A,Type=Float,Description="Estimated ALT Allele Frequencies">
##INFO=<ID=AR2,Number=1,Type=Float,Description="Allelic R-Squared: estimated squared correlation between most probable REF dose and true REF dose">
##INFO=<ID=DR2,Number=1,Type=Float,Description="Dosage R-Squared: estimated squared correlation between estimated REF dose [P(RA) + 2*P(RR)] and true REF dose">
##INFO=<ID=IMP,Number=0,Type=Flag,Description="Imputed marker">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##FORMAT=<ID=DS,Number=A,Type=Float,Description="estimated ALT dose [P(RA) + P(AA)]">
##FORMAT=<ID=GP,Number=G,Type=Float,Description="Estimated Genotype Probability">
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT test1 test2
chr20 60291 . G T . PASS AR2=0.00;DR2=0.00;AF=0.50 GT:DS:GP 0/1:1:0,1,0 0/1:1:0,1,0
chr20 64506 . C T . PASS AR2=0.00;DR2=0.00;AF=0.0051 GT:DS:GP 0/0:0.01:0.99,0.01,0 0/0:0.01:0.99,0.01,0
chr20 68303 . T C . PASS AR2=0.00;DR2=0.00;AF=0.50 GT:DS:GP 0/1:1:0,1,0 0/1:1:0,1,0
chr20 75250 . C T . PASS AR2=0.00;DR2=0.00;AF=0.50 GT:DS:GP 0/1:1:0,1,0 0/1:1:0,1,0
chr20 88108 . T C . PASS AR2=0.00;DR2=0.00;AF=0.87 GT:DS:GP 1/1:1.73:0,0.26,0.73 1/1:1.73:0,0.26,0.73
chr20 159104 . T C . PASS AR2=0.00;DR2=0.00;AF=0.9960 GT:DS:GP 1/1:1.99:0,0.01,0.99 1/1:1.99:0,0.01,0.99
chr20 175269 . T C . PASS AR2=0.00;DR2=0.00;AF=0.9903 GT:DS:GP 1/1:1.98:0,0.02,0.98 1/1:1.98:0,0.02,0.98
chr20 186086 . G A . PASS AR2=0.00;DR2=0.00;AF=0.983 GT:DS:GP 1/1:1.97:0,0.03,0.97 1/1:1.97:0,0.03,0.97
chr20 186183 . G A . PASS AR2=0.00;DR2=0.00;AF=0.983 GT:DS:GP 1/1:1.97:0,0.03,0.97 1/1:1.97:0,0.03,0.97
chr20 198814 . A T . PASS AR2=0.00;DR2=0.00;AF=0.50 GT:DS:GP 0/1:1:0.05,0.89,0.06 0/1:1:0.05,0.89,0.06
chr20 231710 . T G . PASS AR2=0.00;DR2=0.00;AF=0.70 GT:DS:GP 0/1:1.4:0,0.6,0.4 0/1:1.4:0,0.6,0.4
chr20 240166 . T G . PASS AR2=0.00;DR2=0.00;AF=0.49 GT:DS:GP 0/1:0.97:0.03,0.97,0 0/1:0.97:0.03,0.97,0
chr20 240377 . T G . PASS AR2=0.00;DR2=0.00;AF=0.49 GT:DS:GP 0/1:0.97:0.03,0.97,0 0/1:0.97:0.03,0.97,0
chr20 247326 . G A . PASS AR2=0.00;DR2=0.00;AF=0.9964 GT:DS:GP 1/1:1.99:0,0.01,0.99 1/1:1.99:0,0.01,0.99
chr20 248854 . T C . PASS AR2=0.00;DR2=0.00;AF=0.73 GT:DS:GP 0/1:1.47:0,0.53,0.47 0/1:1.47:0,0.53,0.47
chr20 254891 . C T . PASS AR2=0.00;DR2=0.00;AF=0.51 GT:DS:GP 0/1:1.02:0,0.97,0.03 0/1:1.02:0,0.97,0.03
chr20 255081 . G A . PASS AR2=0.00;DR2=0.00;AF=0.85 GT:DS:GP 1/1:1.7:0,0.3,0.7 1/1:1.7:0,0.3,0.7
chr20 260997 . G A . PASS AR2=0.00;DR2=0.00;AF=0.40 GT:DS:GP 0/1:0.8:0.2,0.8,0 0/1:0.8:0.2,0.8,0
chr20 265294 . G T . PASS AR2=0.00;DR2=0.00;AF=0.918 GT:DS:GP 1/1:1.84:0,0.16,0.84 1/1:1.84:0,0.16,0.84
chr20.monovar.vcf
In this vcf file, the high quality SNVs not presented in externtal panel were further genotyped in the cluster level for the study sample. Each column is the cluster identified from scRNA-seq/ATAC-seq clustering.
##fileformat=VCFv4.1
##fileDate=2022-6-19
##source=MonoVar
##FILTER=<ID=LowQual,Description="Low quality">
##INFO=<ID=AC,Number=A,Type=Integer,Description="Allele count in genotypes, for each ALT allele, in the same order as listed">
##INFO=<ID=AF,Number=A,Type=Float,Description="Allele Frequency, for each ALT allele, in the same order as listed">
##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes">
##INFO=<ID=BaseQRankSum,Number=1,Type=Float,Description="Z-score from Wilcoxon rank sum test of Alt Vs. Ref base qualities">
##INFO=<ID=DP,Number=1,Type=Integer,Description="Approximate read depth; some reads may have been filtered">
##INFO=<ID=QD,Number=1,Type=Float,Description="Variant Confidence/Quality by Depth">
##INFO=<ID=SOR,Number=1,Type=Float,Description="Symmetric Odds Ratio of 2x2 contingency table to detect strand bias">
##INFO=<ID=MPR,Number=1,Type=Float,Description="Log Odds Ratio of maximum value of probability of observing non-ref allele to the probability of observing zero non-ref allele">
##INFO=<ID=PSARR,Number=1,Type=Float,Description="Ratio of per-sample Alt allele supporting reads to Ref allele supporting reads">
##FORMAT=<ID=AD,Number=.,Type=Integer,Description="Allelic depths for the ref and alt alleles in the order listed">
##FORMAT=<ID=DP,Number=1,Type=Integer,Description="Approximate read depth (reads with MQ=255 or with bad mates are filtered)">
##FORMAT=<ID=GQ,Number=1,Type=Integer,Description="Genotype Quality">
##FORMAT=<ID=GT,Number=1,Type=String,Description="Genotype">
##FORMAT=<ID=PL,Number=G,Type=Integer,Description="Normalized, Phred-scaled likelihoods for genotypes as defined in the VCF specification">
##reference=file:../example/chr20_2Mb.hg38.fa
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT chr20_0.bam chr20_9.bam chr20_1.bam chr20_10.bam chr20_5.bam chr20_7.bam chr20_3.bam chr20_11.bam chr20_2.bam chr20_16.bam chr20_14.bam chr20_13.bam chr20_8.bam chr20_6.bam chr20_12.bam chr20_4.bam chr20_15.bam chr20_19.bam chr20_18.bam chr20_17.bam
chr20 340478 . T C 34.254262362887154 PASS AC=6;AF=0.18;AN=34;BaseQRankSum=0.43;DP=335;QD=0.19243967619599525;SOR=nan;MPR=85.91;PSARR=0.28 GT:AD:DP:GQ:PL 0/1:76,15:96:6:121,0,1735 0/0:30,3:33:14:0,13,721 0/0:54,7:61:2:3,4,1240 0/0:7,1:8:2:2,4,171 0/1:6,2:8:2:21,0,144 0/0:11,0:11:16:0,13,304 0/1:8,5:15:2:89,0,182 0/0:5,0:5:13:0,12,137 0/1:27,4:33:2:9,1,614 0/0:7,0:7:15:0,13,194 0/0:1,0:1:5:1,7,40 0/0:10,0:10:16:0,13,277 0/0:12,1:14:11:0,11,304 0/1:14,5:19:2:71,0,320 ./. 0/1:3,3:7:2:57,0,71 0/0:6,0:6:14:0,12,171 0/0:1,0:1:5:1,7,40 ./. ./. <10001010100001X100XX>
chr20 410526 . C T 323 PASS AC=20;AF=0.53;AN=38;BaseQRankSum=-0.24;DP=355;QD=0.9098591549295775;SOR=nan;MPR=500.13;PSARR=0.86 GT:AD:DP:GQ:PL 0/1:49,31:83:3:585,0,1064 0/1:2,4:6:3:90,0,38 0/1:34,30:64:3:586,0,714 0/1:11,10:21:3:204,0,232 0/1:23,19:42:3:380,0,488 0/1:4,5:9:3:107,0,82 0/1:10,12:22:3:254,0,199 1/1:0,4:4:3:106,5,1 0/1:10,12:22:3:254,0,202 0/1:2,4:6:3:90,0,38 0/1:2,4:7:3:90,0,38 0/1:4,5:9:3:107,0,66 0/1:5,4:10:3:73,0,109 0/1:6,3:9:3:38,0,135 0/1:8,2:10:3:25,0,169 0/1:13,7:22:3:127,0,288 0/1:3,3:6:3:63,0,65 0/1:0,1:1:2:28,3,4 ./. 0/1:1,1:2:3:22,0,24 <111111121111111111X1>
chr20 453027 . T C 37.74965741923828 PASS AC=8;AF=0.21;AN=38;BaseQRankSum=-0.15;DP=489;QD=0.1906548354506984;SOR=nan;MPR=94.06;PSARR=0.15 GT:AD:DP:GQ:PL 0/1:70,10:83:8:18,0,1620 0/0:26,0:28:11:0,13,698 0/1:35,11:48:3:134,0,796 0/1:18,6:24:3:83,0,415 0/0:56,3:62:11:0,13,1379 0/0:10,0:10:11:0,13,277 0/0:22,1:24:11:0,13,549 0/1:10,5:15:3:84,0,210 0/0:37,3:41:11:0,13,918 0/0:25,0:25:11:0,13,657 0/1:8,3:12:3:42,0,188 0/0:6,0:6:10:0,12,171 0/0:36,0:37:11:0,13,964 0/0:24,1:27:11:0,13,602 0/1:6,1:7:2:4,2,149 0/0:24,3:29:4:1,6,571 0/1:4,1:5:2:8,1,101 0/0:2,0:2:6:1,8,65 ./. 0/1:1,2:4:3:40,0,26 <101100010010001010X1>
chr20 484887 . T C 54.16552858770515 PASS AC=6;AF=0.15;AN=40;BaseQRankSum=-0.18;DP=620;QD=0.19840852962529357;SOR=nan;MPR=131.60;PSARR=0.21 GT:AD:DP:GQ:PL 0/1:80,27:145:9:384,0,1808 0/0:21,2:33:9:0,12,508 0/0:46,5:76:9:0,12,1092 0/0:5,0:26:9:0,12,144 0/1:8,2:56:3:19,0,191 0/1:3,2:20:3:34,0,73 0/0:24,1:49:9:0,12,622 0/0:3,0:9:7:0,10,91 0/0:28,2:85:9:0,12,701 0/1:1,1:9:3:17,0,25 0/0:3,0:10:7:0,10,91 0/0:0,0:7:3:2,5,14 0/1:3,3:22:3:58,0,70 0/0:8,0:26:9:0,12,223 0/0:4,0:9:8:0,11,117 0/1:9,4:21:3:63,0,208 0/0:2,0:2:6:1,8,65 0/0:0,0:3:3:2,5,14 0/0:1,0:3:4:1,7,39 0/0:0,0:9:3:2,5,14 <10001100010010010000>
chr20 646770 . A G 323 PASS AC=19;AF=0.47;AN=40;BaseQRankSum=-0.51;DP=327;QD=1.0;SOR=nan;MPR=516.72;PSARR=0.81 GT:AD:DP:GQ:PL 0/1:24,28:53:3:577,0,465 0/1:1,2:3:3:45,0,21 0/1:13,12:25:3:245,0,274 0/1:20,19:39:3:373,0,414 0/1:35,20:55:3:356,0,760 0/1:9,8:17:3:162,0,191 0/1:11,9:21:3:180,0,236 0/1:5,4:9:3:64,0,109 0/1:7,10:17:3:216,0,138 0/1:8,1:9:2:4,2,192 0/1:8,1:9:2:4,2,188 0/1:5,1:6:3:10,0,119 0/1:7,2:9:3:27,0,162 0/1:12,8:20:3:138,0,262 0/1:2,4:6:3:89,0,39 0/1:5,4:10:3:80,0,106 0/1:3,4:7:3:86,0,62 0/1:2,2:4:3:26,0,45 0/1:2,2:4:3:42,0,45 0/0:4,0:4:5:1,7,109 <11111111111111111110>
chr20 1143853 . A G 19.800259769261785 PASS AC=6;AF=0.16;AN=38;BaseQRankSum=0.36;DP=343;QD=0.21758527218968995;SOR=nan;MPR=52.52;PSARR=0.15 GT:AD:DP:GQ:PL 0/0:74,10:87:6:1,6,1663 0/0:26,1:27:11:0,13,652 0/1:18,5:24:3:55,0,407 0/0:17,1:24:11:0,13,426 0/1:12,3:19:3:30,0,279 0/0:13,0:14:11:0,13,334 0/1:5,3:9:3:51,0,102 0/0:10,0:11:11:0,13,262 0/0:37,0:38:11:0,13,948 0/1:11,3:16:3:30,0,220 0/0:1,0:1:5:1,7,40 0/1:10,3:15:3:32,0,216 0/0:16,2:19:4:1,6,386 0/0:12,0:12:11:0,13,327 0/1:5,2:8:3:27,0,117 0/0:8,1:9:3:1,5,189 0/0:6,0:6:10:0,12,171 0/0:2,0:2:6:1,9,62 ./. 0/0:1,0:2:5:1,7,40 <001010100101001000X0>
chr20 1288715 . A G 11.405507006226141 PASS AC=5;AF=0.18;AN=28;BaseQRankSum=0.25;DP=337;QD=0.20366976796832395;SOR=nan;MPR=33.17;PSARR=0.16 GT:AD:DP:GQ:PL 0/0:117,15:134:7:1,7,3240 0/0:17,1:18:3240:0,14,432 0/0:79,5:86:3240:0,14,1950 0/1:4,1:6:2:7,1,99 0/0:9,0:9:3240:0,14,254 0/0:7,0:10:3240:0,14,201 0/1:21,6:29:2:69,0,464 ./. 0/1:12,3:15:2:29,0,268 0/1:3,1:4:2:9,1,75 ./. 0/1:0,1:2:1:19,1,9 0/0:8,0:8:3240:0,14,220 0/0:7,0:7:3240:0,14,201 0/0:6,1:7:1:3,3,152 ./. ./. ./. 0/0:2,0:2:9:0,10,69 ./. <0001001X11X1000XXX0X>
chr20 1435877 . A G 323 PASS AC=19;AF=0.50;AN=38;BaseQRankSum=0.08;DP=549;QD=0.5883424408014571;SOR=0.00;MPR=357.70;PSARR=1.65 GT:AD:DP:GQ:PL 0/1:19,33:52:3:721,0,348 0/1:10,17:27:3:356,0,188 0/1:13,21:34:3:453,0,239 0/1:15,33:48:3:722,0,254 0/1:19,18:37:3:364,0,393 0/1:7,20:27:3:435,0,105 0/1:18,29:47:3:614,0,337 0/1:5,12:17:3:265,0,85 0/1:16,44:60:3:986,0,248 0/1:25,54:79:3:1194,0,398 0/1:6,10:16:3:219,0,115 0/1:10,5:15:3:85,0,220 0/1:3,19:22:3:420,0,17 0/1:14,3:17:3:30,0,324 0/1:4,3:7:3:60,0,89 0/1:11,4:15:3:62,0,250 0/1:3,7:10:3:157,0,53 0/1:7,6:13:3:118,0,150 ./. 0/1:2,4:6:3:89,0,38 <111111111111111111X1>
chr20 1443658 . G A 323 PASS AC=20;AF=0.50;AN=40;BaseQRankSum=-0.63;DP=1210;QD=0.26694214876033057;SOR=1.17;MPR=9.95;PSARR=0.84 GT:AD:DP:GQ:PL 0/1:98,99:197:3:3240,0,3240 0/1:20,26:46:3:3240,0,3240 0/1:60,49:109:3:3240,0,3240 0/1:37,31:68:3:3240,0,3240 0/1:74,58:132:3:3240,0,3240 0/1:23,26:49:3:3240,0,3240 0/1:66,47:113:3:3240,0,3240 0/1:14,18:32:3:3240,0,3240 0/1:61,51:112:3:3240,0,3240 0/1:33,23:56:3:3240,0,3240 0/1:12,7:19:3:3240,0,3240 0/1:18,15:33:3:3240,0,3240 0/1:33,28:61:3:3240,0,3240 0/1:30,35:65:3:3240,0,3240 0/1:14,14:28:3:3240,0,3240 0/1:34,15:49:3:3240,0,3240 0/1:7,3:10:3:3240,0,3240 0/1:9,4:13:3:3240,0,3240 0/1:5,1:6:3:3240,0,3240 0/1:8,4:12:3:3240,0,3240 <11111111111111111111>
chr20 1462725 . A G 323 PASS AC=20;AF=0.50;AN=40;BaseQRankSum=0.59;DP=2282;QD=0.14154250657318143;SOR=1.43;MPR=718.09;PSARR=0.89 GT:AD:DP:GQ:PL 0/1:371,284:661:0:811,0,1223 0/1:94,107:201:0:1159,0,894 0/1:212,130:342:0:678,0,1368 0/1:53,60:113:0:1019,0,1027 0/1:97,58:156:0:757,0,1281 0/1:36,59:95:0:1277,0,650 0/1:54,64:118:0:1129,0,920 0/1:20,14:34:0:269,0,424 0/1:76,115:191:0:1524,0,502 0/1:16,16:32:0:324,0,312 0/1:23,30:53:0:633,0,435 0/1:17,15:32:0:298,0,354 0/1:29,34:64:0:702,0,580 0/1:27,24:52:0:483,0,563 0/1:10,18:28:0:393,0,181 0/1:36,16:52:0:268,0,783 0/1:6,14:20:0:311,0,98 0/1:1,1:2:0:3240,0,3240 0/1:13,10:23:0:192,0,256 0/1:9,4:13:0:66,0,199 <11111111111111111111>
chr20 1763453 . T C 323 PASS AC=16;AF=0.47;AN=34;BaseQRankSum=1.06;DP=1111;QD=0.2928377153218495;SOR=inf;MPR=352.78;PSARR=1.02 GT:AD:DP:GQ:PL 0/1:275,204:479:3:884,0,1157 0/1:29,46:75:3:996,0,546 0/1:132,132:264:3:1309,0,662 0/0:3,0:3:6:1,7,85 0/1:18,3:21:3:17,0,384 1/1:0,3:3:1:77,4,2 0/1:28,8:36:3:104,0,633 0/1:8,10:18:3:211,0,159 0/1:1,2:3:3:44,0,22 0/1:2,1:3:3:18,0,49 0/1:7,7:14:3:144,0,132 0/0:1,0:1:3:2,4,33 1/1:0,4:4:2:103,4,2 0/1:21,16:37:3:314,0,451 0/0:4,0:4:8:1,8,111 0/1:33,41:74:3:867,0,651 0/1:40,30:72:3:571,0,841 ./. ./. ./. <11101211111021011XXX>
chr20 1799734 . C A 323 PASS AC=13;AF=0.50;AN=26;BaseQRankSum=-0.71;DP=459;QD=0.7146017699115044;SOR=nan;MPR=424.72;PSARR=1.01 GT:AD:DP:GQ:PL 0/1:93,78:171:2:1529,0,3240 0/1:11,10:21:2:201,0,235 0/1:53,52:108:2:1032,0,1079 0/1:5,4:9:2:74,0,112 0/0:3,0:3:3240:1,8,87 ./. 0/1:8,8:16:2:155,0,171 1/1:0,4:4:1:100,3,3 1/1:0,4:4:1:100,3,3 ./. 0/1:0,1:1:1:24,1,6 0/0:4,0:4:3240:1,9,114 ./. 0/1:7,6:13:2:119,0,153 ./. 0/1:26,31:57:2:635,0,526 0/1:36,12:48:2:146,0,802 ./. ./. ./. <11110X122X10X1X11XXX>
chr20 1893313 . G A 323 PASS AC=16;AF=0.47;AN=34;BaseQRankSum=-0.14;DP=459;QD=0.7067833698030634;SOR=nan;MPR=475.42;PSARR=0.94 GT:AD:DP:GQ:PL 0/1:121,70:218:3:1247,0,3240 0/1:7,5:15:3:97,0,154 0/1:46,43:112:3:872,0,942 0/1:2,3:5:3:62,0,42 0/1:4,3:7:3:59,0,89 0/1:3,1:4:3:15,0,72 0/1:5,19:28:3:429,0,65 0/1:9,4:13:3:68,0,200 0/1:0,1:1:2:27,2,4 0/1:0,1:1:2:23,2,4 1/1:0,3:8:2:78,4,2 0/0:1,0:1:3:2,4,32 0/0:1,0:1:3:2,4,32 0/1:2,5:15:3:112,0,36 ./. 0/1:9,14:25:3:303,0,174 0/1:1,2:4:3:41,0,21 ./. ./. 0/1:0,1:1:2:27,2,4 <11111111112001X11XX1>
chr20 1900266 . G A 323 PASS AC=14;AF=0.44;AN=32;BaseQRankSum=0.37;DP=539;QD=0.6345776031434185;SOR=0.85;MPR=396.97;PSARR=0.90 GT:AD:DP:GQ:PL 0/1:100,103:211:3:3240,0,3240 0/1:34,13:48:3:203,0,736 0/1:64,70:137:3:1445,0,1291 0/0:11,1:17:8:1,8,272 0/1:5,5:10:3:102,0,108 ./. 0/1:16,9:28:3:163,0,351 0/1:3,4:8:3:84,0,64 0/0:10,0:11:3240:0,10,255 1/1:0,5:5:2:112,4,2 ./. ./. 0/1:1,3:4:3:67,0,20 0/1:1,2:7:3:43,0,23 0/1:0,1:1:2:25,2,5 0/1:26,11:38:3:177,0,581 0/1:3,7:11:3:151,0,55 ./. 0/0:2,0:2:5:1,6,60 0/1:0,0:1:2:4,3,10 <11101X1102XX11111X01>
-
where to download 1KG3 reference panel (hg38) http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20201028_3202_phased/
-
how to perform downstream PCA-based projection or admixture analysis
PCA-based projection analysis can be peformed using LASER 2.0