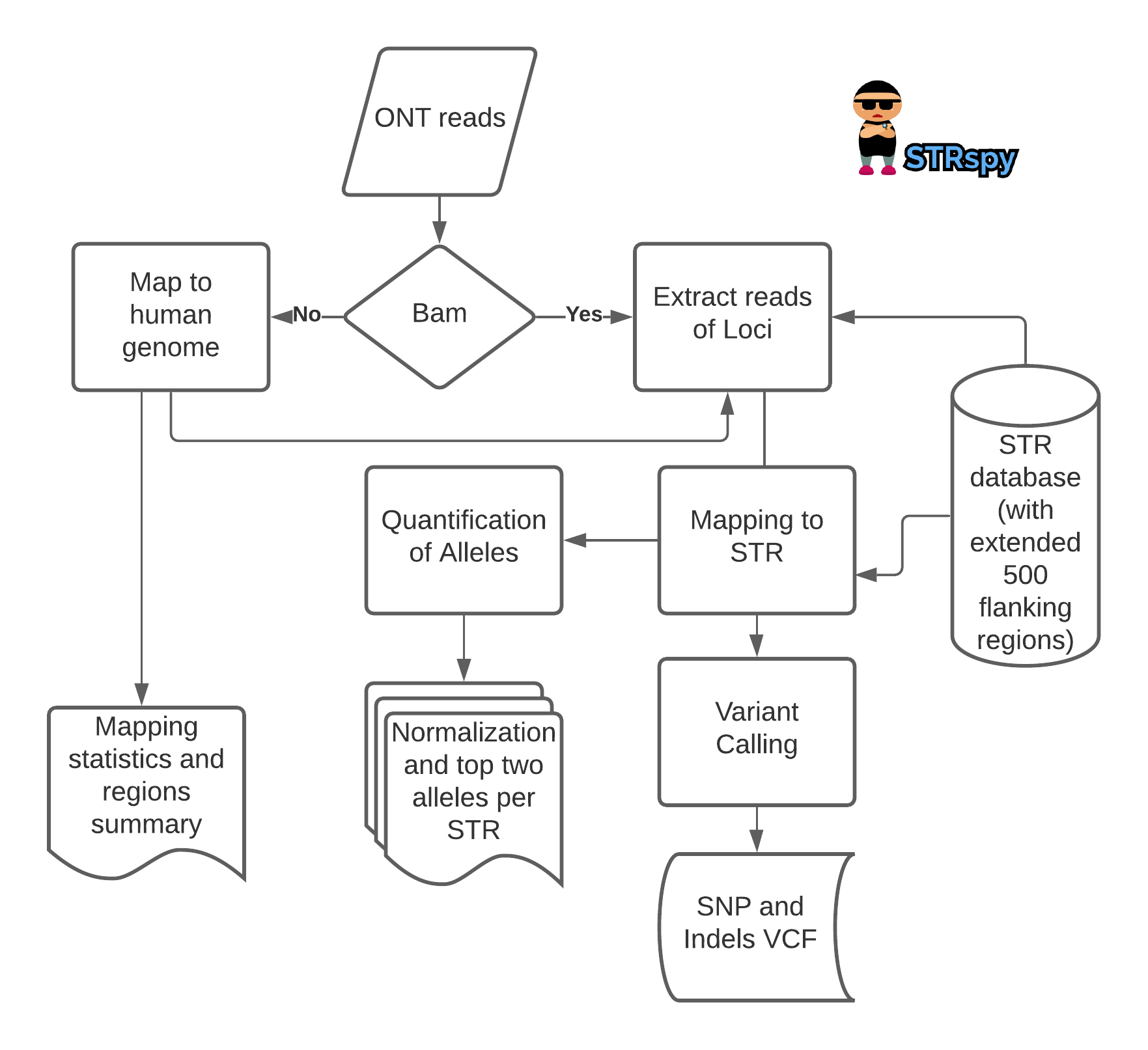
STRspy: a novel alignment and quantification-based state-of-the-art method, short tandem repeat (STR) detection calling tool designed specifically for long-read sequencing reads such as from Oxford nanopore technology (ONT) and PacBio.
DNA evidence has long been considered the gold standard for human identification in forensic investigations. Most often, DNA typing exploits the high variability of short tandem repeat (STR) sequences to differentiate between individuals at the genetic level. Comparison of STR profiles can be used for human identification in a wide range of forensic cases including homicides, sexual assaults, missing persons, and mass disaster victims. The number of contiguous repeat units present at a given microsatellite locus varies significantly among individuals, and thus make them useful for human identification purposes. Here, we are presents a complete pipeline i.e STRspy to identify STRs in a long read samples i.e. Oxford nanopore sequencing reads and Pacbio reads.
- Input either fastq (raw reads usually from ONT) or bam (pre-aligned reads usually froom PacBio)
- Reports raw counts of allele along with its Normalized counts by its maximum value
- Find top two significant Alleles (filtering threshold set by user such as 0.4)
- Detects Small variants such as SNP and Indels
- Reports mapping summary and STR region of overlaps
- Stutters analysis for simple motifs of STRs
1.1 Install Miniconda
Download Miniconda installer from here: https://docs.conda.io/en/latest/miniconda.html and Install it to your laptop or server.
wget https://repo.anaconda.com/miniconda/Miniconda3-py39_4.9.2-Linux-x86_64.sh
bash Miniconda3-py39_4.9.2-Linux-x86_64.sh
Follow the instructions directed by the miniconda script
1.2 Install STRspy
STRspy includes the installation of the following third-party software before it can be used.
gnu parallel >=20210222
samtools >=v1.12
bedtools >=v2.30.0
minimap2 >=v2.18-r1015
xatlas >=v0.2.1
git clone git@github.com:unique379r/strspy.git
cd strspy
bash setup/STRspy_setup.sh
conda activate strspy_env
bash setup/MakeToolConfig.sh
mv UserToolsConfig.txt config/
conda deactivate
Modify the configfiles describing your data config/InputConfig.txt
cd strspy
bash ./STRspy_run_v1.0.sh -h
USAGE: bash ./STRspy_run_v1.0.sh config/InputConfig.txt config/ToolsConfig.txt
The testset is provided testset.tar.gz with the package for the quick start, however pre-computed results test_results.tar.gz are also avaiable for the reproducibility purposes. The test data should finish less than 12 Sec (via simple terminal use) to generate the results.
tar -xvf testset.tar.gz
Compare your test results with pre-computed outputs here
tar -xvf test_results.tar.gz
INPUT_DIR : A dir must have either fastq (Oxford nanopore genomic reads) or bam (aligned genomic reads such as from PacBio)
INPUT_BAM : Given inputs are bam or fastq (yes or no)
READ_TYPE : Sequencing Technology (ont or pb)
STR_FASTA : A dir contains Fasta files for each STR region of interest [assimung it has flanking regions (+/-) of 500bp]
STR_BED : A dir contains Bed files for each STR region of interest [assimung it has flanking regions (+/-) of 500bp]
GENOME_FASTA: Genome fasta (hg19/hg38) must provide in case of fastq input.
REGION_BED : All STr bed has to concatnated inot single bed file to calculate the coverage of it from the aligment sample file.
NORM_CUTOFF : A normalization threshold is required to select top two allles of a STR
OUTPUT_DIR : A empty directory to write the results
BEDTOOLS = ../user/path/bedtools
MINIMAP = ../user/path/minimap2
SAMTOOLS = ../user/path/samtools
XATLAS = ../user/path/xatlas
PARALLEL = ../user/path/parallel
One may encounter with a bug that using wrapper (STRspy_run_v1.0.sh), STRspy parallel version may not able to communicate properly with "gnu parallel" and exit the workfow without mapping or further steps of the analysis. Solution to this, the user may either run the script directly from scripts/STRspy_v1.0.sh in the STRspy dir or modify the STRspy_run_v1.0.sh script and allow the nested loop version of the workflow, but bear in mind that this is a little slower than the parallel version.
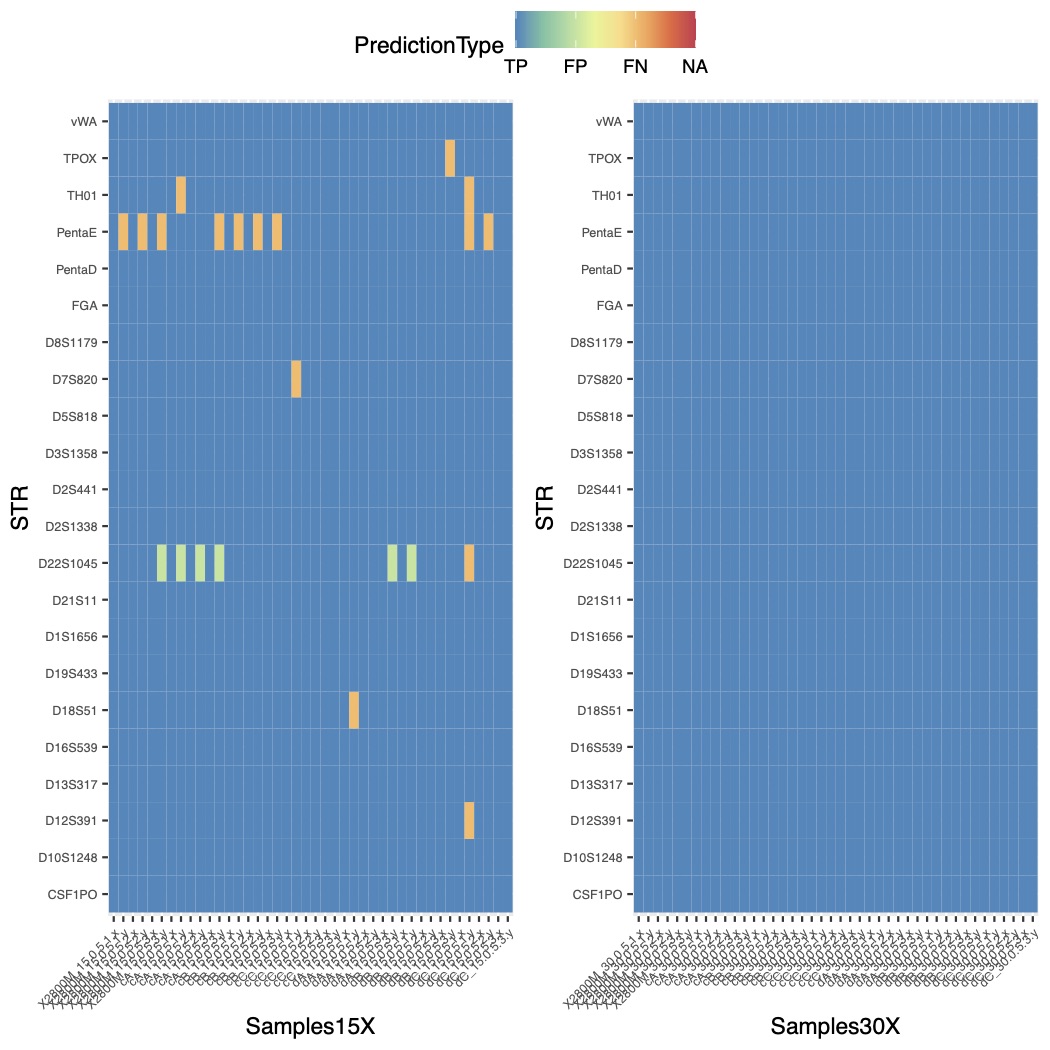
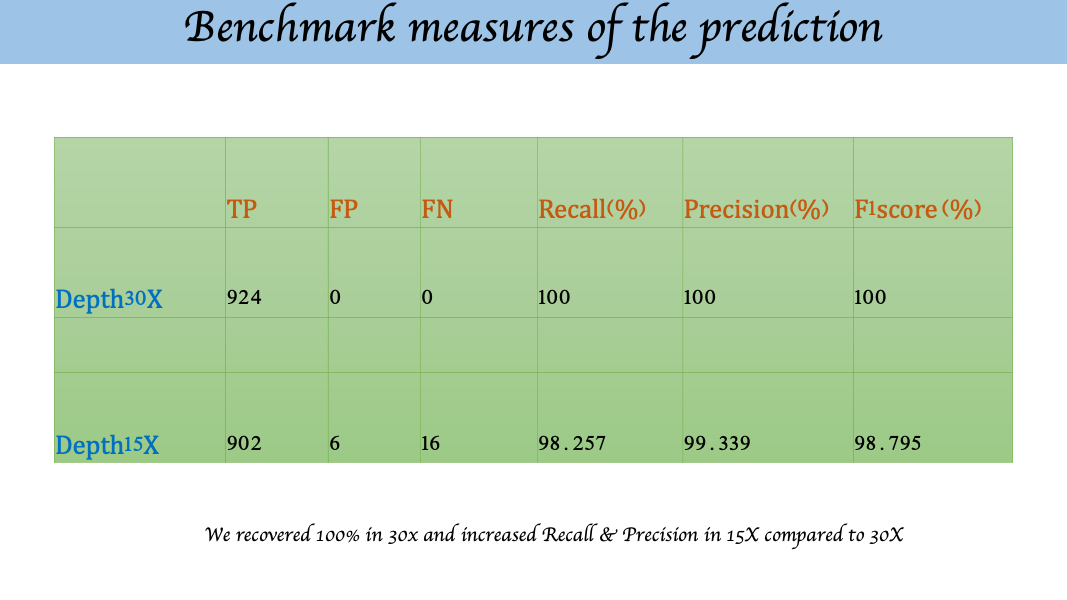
STRspy has been evaluated on 2 datasets including 30cycle and 15cycle of the ONT reads. Please have a look plots bellow the benchmarking of the datasets we used. For more details please have a look our paper Courtney L. Halla*, Rupesh K. Kesharwani et al. 2021; Accurate profiling of forensic autosomal STRs using the Oxford Nanopore Technologies MinION device; Forensic Science International: Genetics.
Aaron R. Quinlan, Ira M. Hall, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, Volume 26, Issue 6, 15 March 2010, Pages 841–842, https://doi.org/10.1093/bioinformatics/btq033
Li, H. (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34:3094-3100. https://doi:10.1093/bioinformatics/bty191
Heng Li, Bob Handsaker, Alec Wysoker, Tim Fennell, Jue Ruan, Nils Homer, Gabor Marth, Goncalo Abecasis, Richard Durbin, 1000 Genome Project Data Processing Subgroup, The Sequence Alignment/Map format and SAMtools, Bioinformatics, Volume 25, Issue 16, 15 August 2009, Pages 2078–2079, https://doi.org/10.1093/bioinformatics/btp352
Jesse Farek, Daniel Hughes, Adam Mansfield, Olga Krasheninina et al (2018). xAtlas: Scalable small variant calling across heterogeneous next-generation sequencing experiments. bioRxiv; doi: https://doi.org/10.1101/295071
O. Tange (2018): GNU Parallel 2018, March 2018, https://doi.org/10.5281/zenodo.1146014.
bioinforupesh200 DOT au AT gmail DOT com
rupesh DOT kesharwani AT bcm DOT edu