This repository includes the code required to reproduce the experiments and figures in the paper:
Yushan Huang, Yuchen Zhao, Capstick Alexander, Francesca Palermo, Hamed Haddadi, Payam Barnaghi. "Analyzing entropy features in time-series data for pattern recognition in neurological conditions." Accepted to Artificial Intelligence in Medicine, Elsevier. Paper, Code
To get started and download all dependencies, run:
pip install -r requirements.txt
The Minder dataset is privacy. To apply and download the dataset, please contact with Prof. Payam Barnaghi.
Generate entropy features, shown as EntropyFeatures.
Entropy features for the Minder dataset includes:
Entropy of Markov chains: ./MINDER/EntropyFeatures/activity_daytime_per_week_mk_entropy.ipynb, ./MINDER/EntropyFeatures/activity_night_per_week_mk_entropy.ipynb
Entropy rate of Markov chains: ./MINDER/EntropyFeatures/activity_daytime_per_week_mk_entropy_rate.ipynb, ./MINDER/EntropyFeatures/activity_night_per_week_mk_entropy_rate.ipynb
Entropy production of Markov chains: ./MINDER/EntropyFeatures/activity_daytime_per_week_mk_entropy_production.ipynb, ./MINDER/EntropyFeatures/activity_night_per_week_mk_entropy_production.ipynb
Von Neumann Entropy of Markov chains (activity frequency): ./MINDER/EntropyFeatures/activity_daytime_per_week_mk_vn_entropy_frequency.ipynb, ./MINDER/EntropyFeatures/activity_night_per_week_mk_vn_entropy_frequency.ipynb
Von Neumann Entropy of Markov chains (activity duration): ./MINDER/EntropyFeatures/activity_daytime_per_week_mk_vn_entropy_duration.ipynb, ./MINDER/EntropyFeatures/activity_night_per_week_mk_vn_entropy_duration.ipynb
The baseline features: ./MINDER/EntropyFeatures/activity_daytime_night_per_week_frequency.ipynb
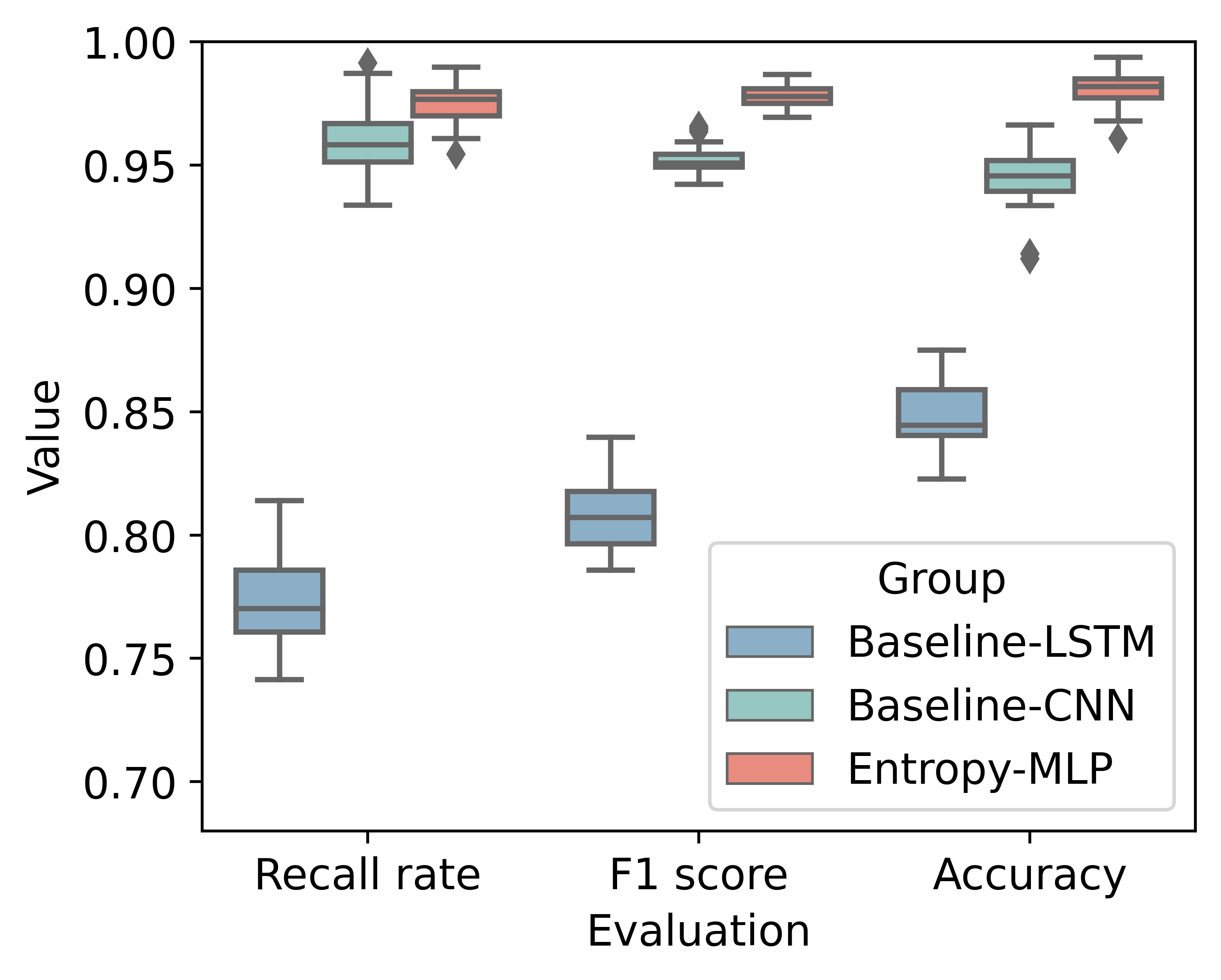
The evaluation results are:
This repository includes the code for Epileptic Seizure Recognition Dataset (ESRD).
Download the orginal data[1].
The code is in ./ESRD/plot_raw.ipynb.
Generate entropy features, shown as ./ESRD/generate_entropy.ipynb.
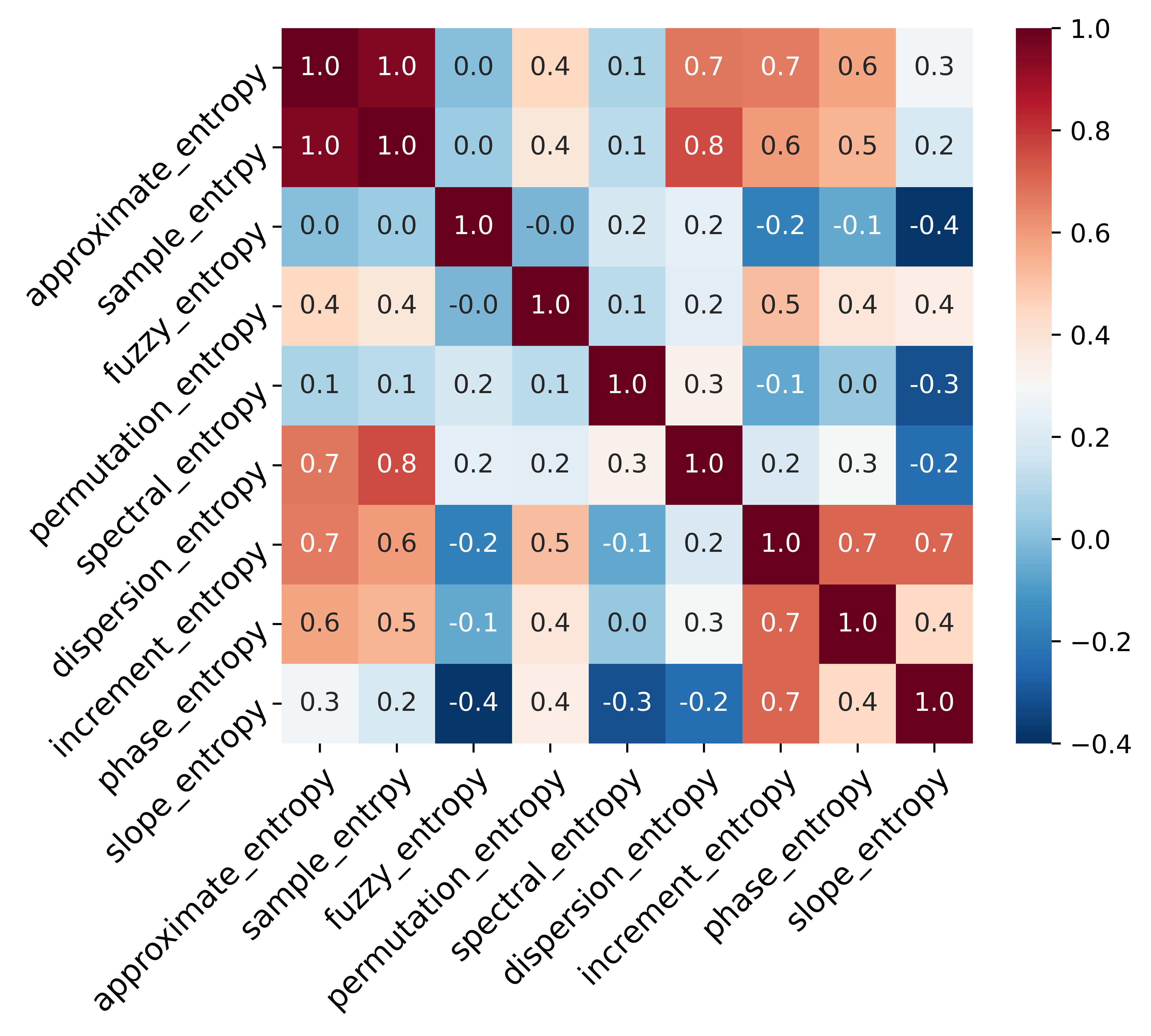
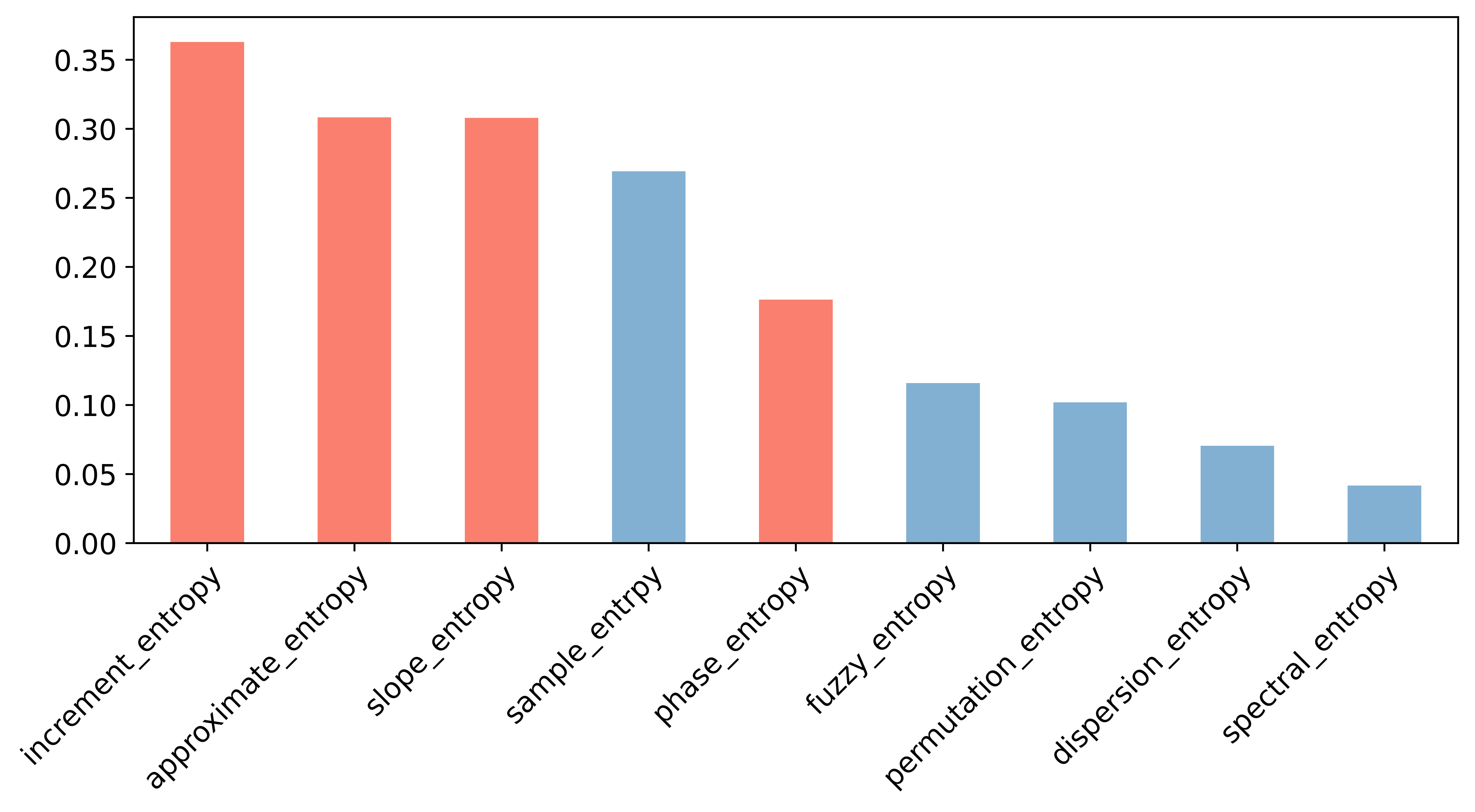
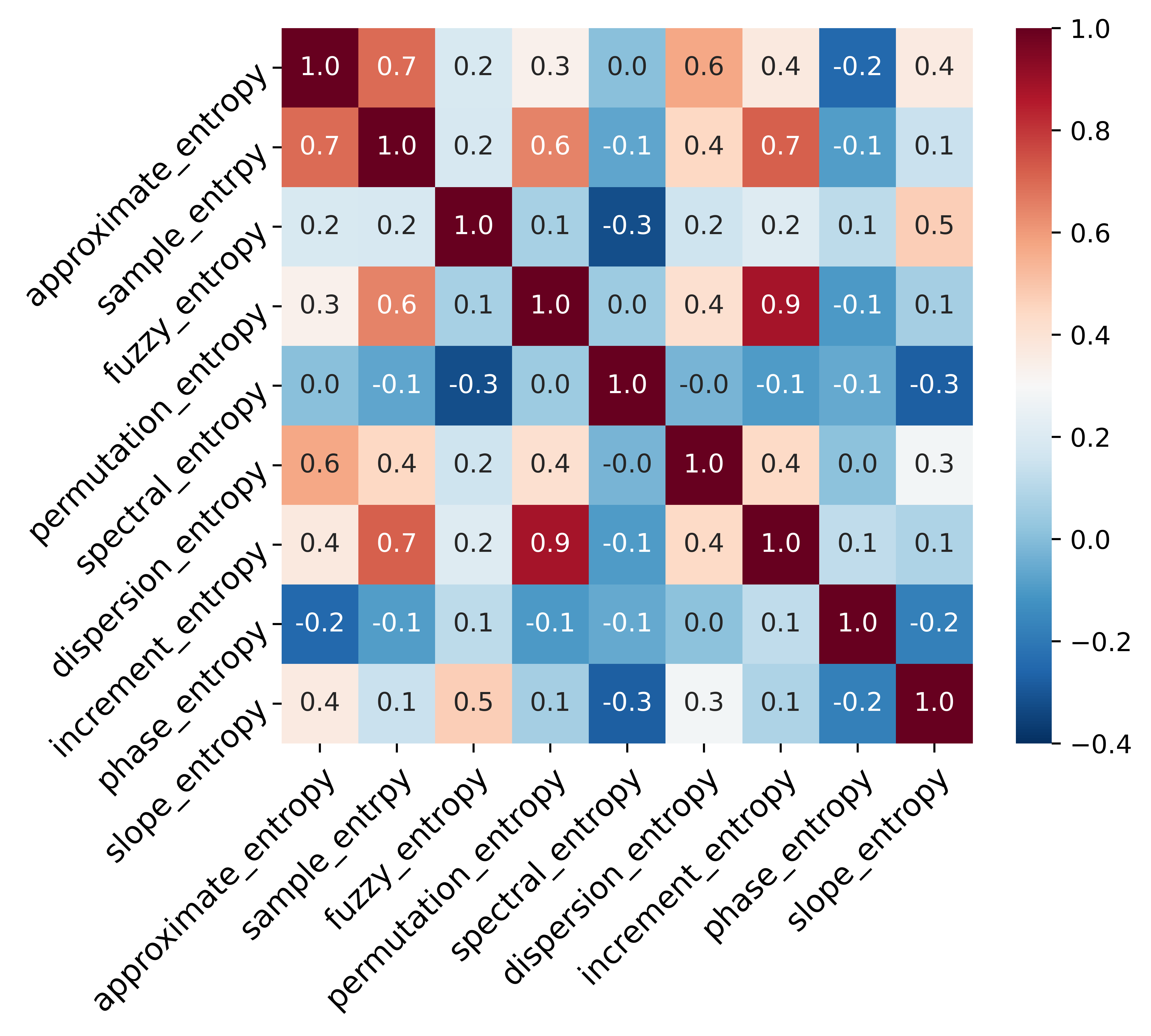
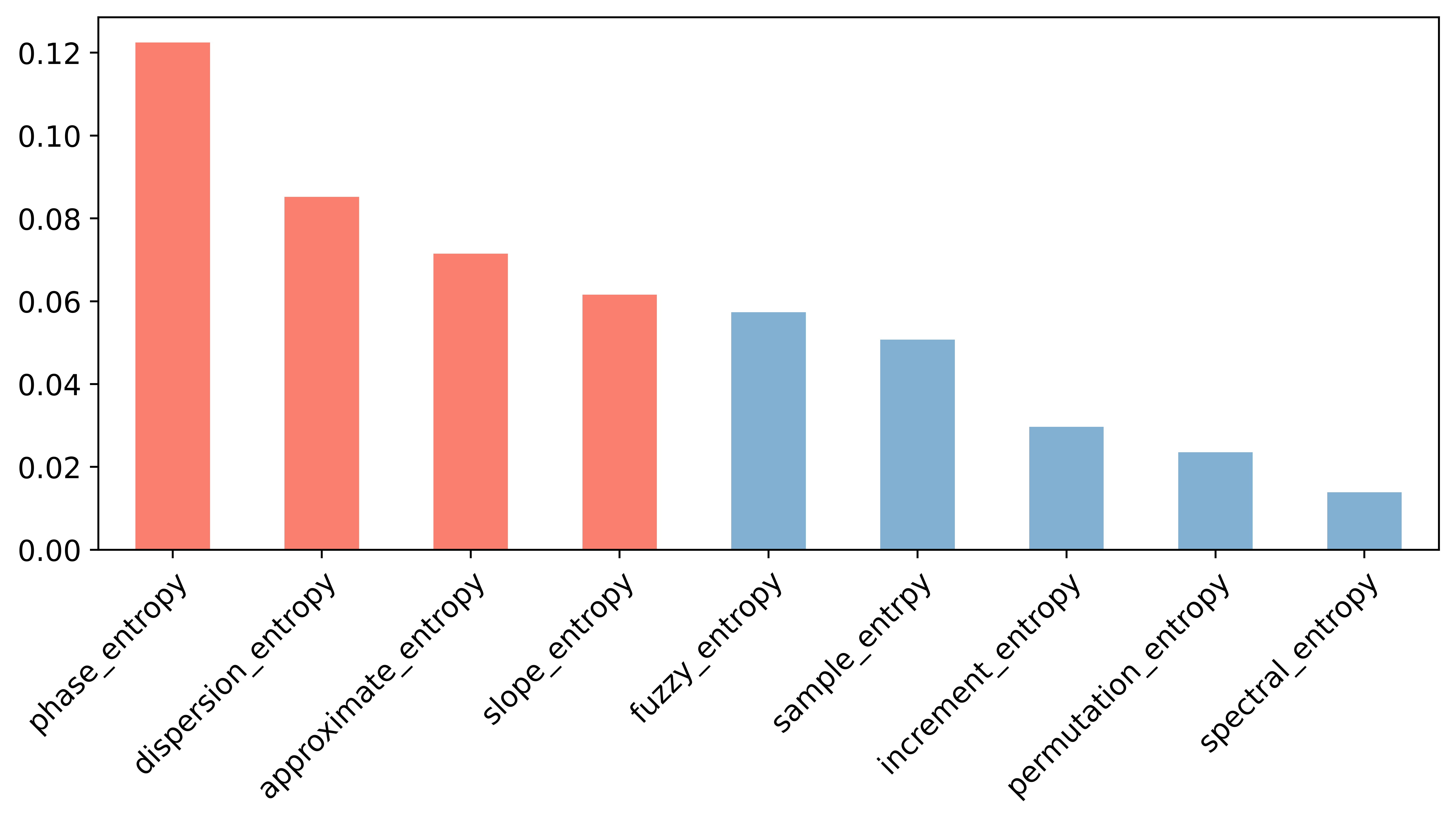
Select entropy features by Pearson relationship matrix and mutual information.
The code is shown in ./ESRD/model_baseline_CNN.ipynb.
The code is shown in ./ESRD/model_baseline_LSTM.ipynb.
The code is shown in ./ESRD/network_pytorch.
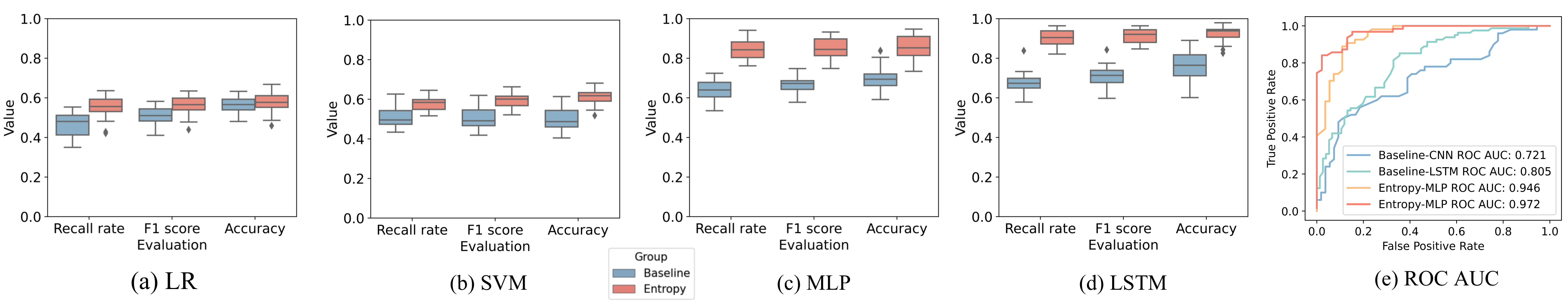
The evaluation results are:
[1]Andrzejak, R.G., Lehnertz, K., Mormann, F., Rieke, C., David, P. and Elger, C.E., 2001. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Physical Review E, 64(6), p.061907.
This repository includes the code for PTB Diagnostic ECG Database.
Download the orginal data[2].
Download the pre-processed data.
Here we utilize the pre-processed data.
For the normal participants:
For the abnormal participants:
The code is in ./PTBDB/generate_entropy.ipynb.
Generate entropy features, shown as ./PTBDB/generate_entropy.ipynb.
Select entropy features by Pearson relationship matrix and mutual information.
The code is shown in ./PTBDB/model_baseline_CNN.ipynb.
The code is shown in ./PTBDB/model_baseline_MLP.ipynb.
The code is shown in ./PTBDB/network_pytorch.
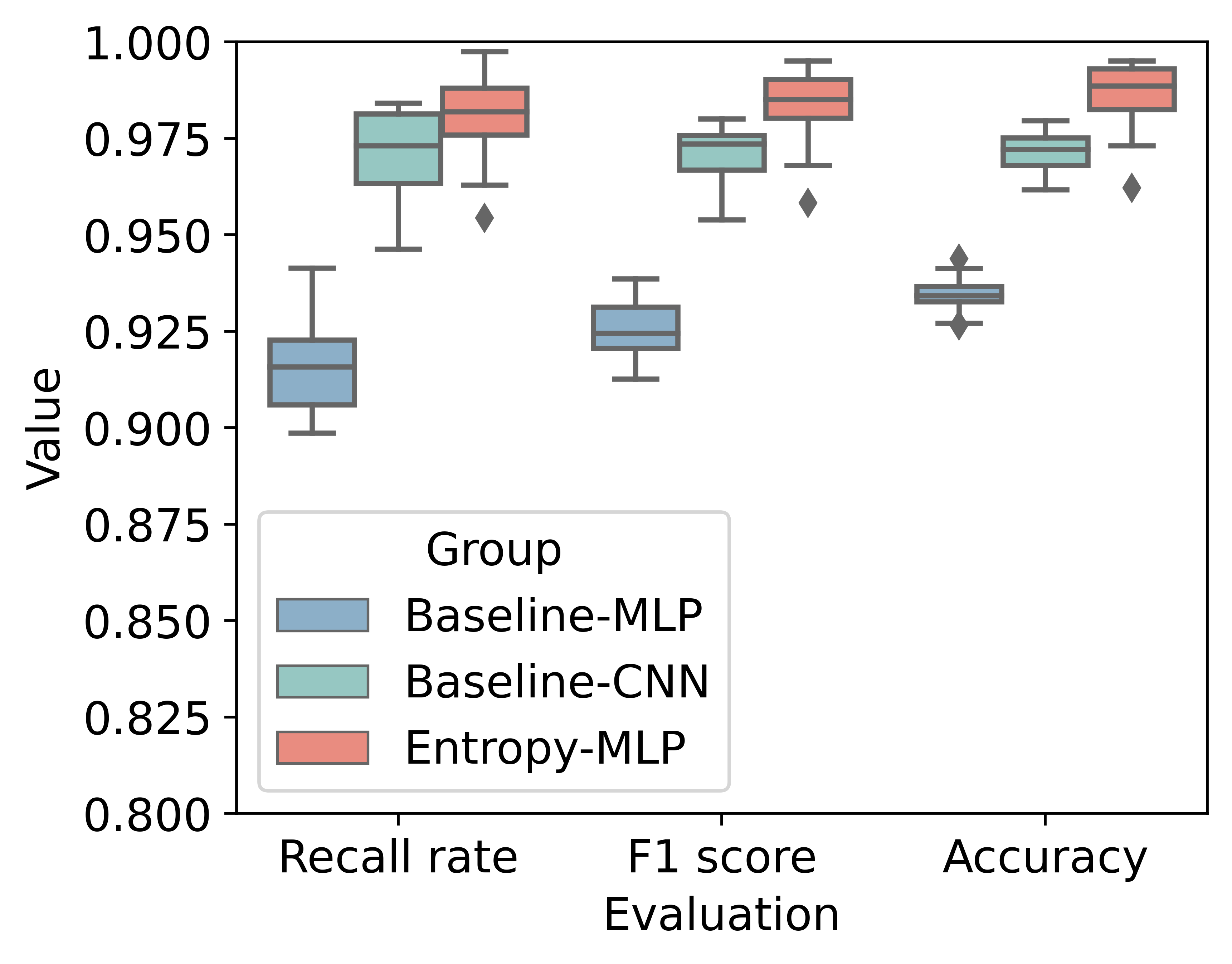
The evaluation results are:
The code is shown in ./PTBDB/plot_result.ipynb.
[1]Andrzejak, R.G., Lehnertz, K., Mormann, F., Rieke, C., David, P. and Elger, C.E., 2001. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Physical Review E, 64(6), p.061907.
[2]Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215 - e220.