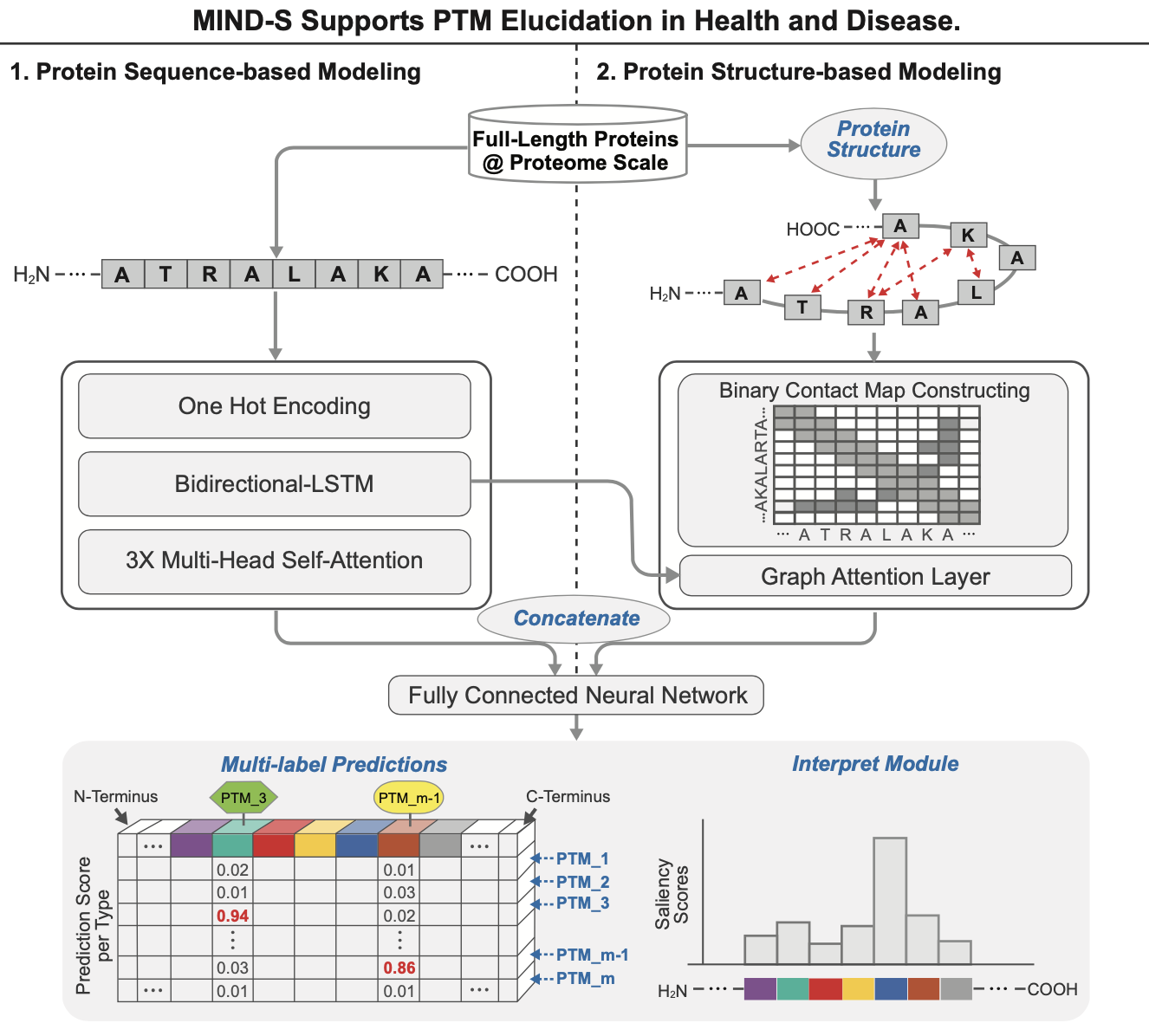
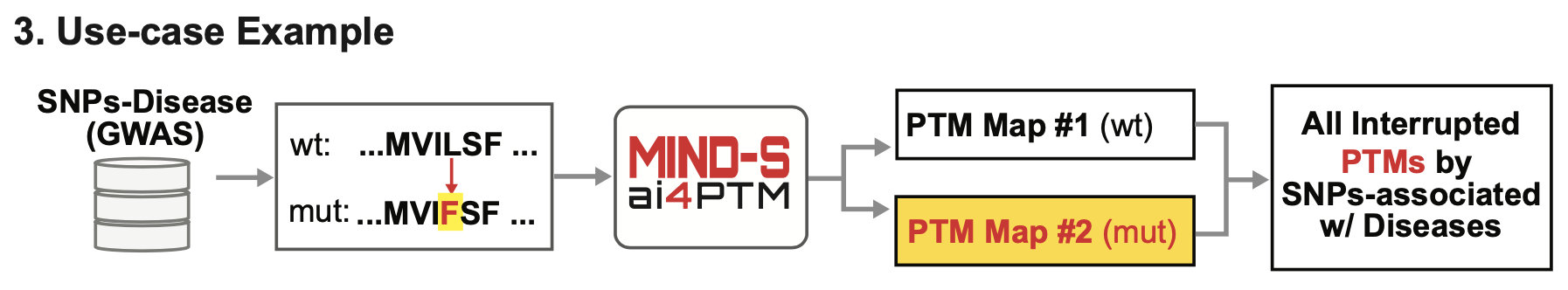
MIND-S (Multilabel INterpretable Deep learning method for PTM prediction) is a deep learning tool for making PTM predictions based on protein sequence, or protein structure. MIND-S features interpretability of the model through evaluating the importance for each input residual to identify the important residual for making a prediction. MIND-S can also be utilized as a tool for evaluating effects of mutations (e.g. SNPs) that eventually affect protein sequence. By comparing the PTM predictions between wildtype and mutant protein, MIND-S can give hint on whether the mutation will affect PTMs.
Install MIND:
git clone https://github.com/yuyanislearning/MIND.git
cd MINDWe suggest building environment via docker or conda or using platforms with tensorflow2 installed.
We provide a Dockerfile which directly sets up the tensorflow and relevant package. More information about docker can be found here. The prerequisite for using the docker can be found here. After prerequisite is satisfied, you can use the dockerfile here Run the following to build a docker image
cd docker_buildmv [path to Downloaded dockerfile]/Dockerfile ./You need to replace [path to Downloaded dockerfile] with your file path of the Dockerfile you downloaded.
docker build -t yuyanislearning/mind:1.0 .Then run the following to run a docker container
docker run --gpus all -it --rm -v [Path to your working directory, need to contain MIND]:/workspace yuyanislearning/mind:1.0Follow the instruction to install tensorflow2.
Install required packages:
pip install -r requirementMIND allows batch predictions for multiple proteins. A fasta files contains all protein sequence can be used as the input with run the following code. A json file with ptm information (uid_site_PtmType) and prediction scores will be return. An example code using protein Q5S007 fasta sequence is shown below:
mkdir temp
python batch_predict.py \
--pretrain_name saved_model/MIND_fifteenfold \
--data_path sample/Q5S007.fa\
--res_path temp \
--n_fold 15 MIND supports interpretation for individual PTM prediction. The fasta file of the protein interested should be provided and the ptm site and ptm type should also be provided. A list of supported ptm types are list here: 'Hydro_K','Hydro_P','Methy_K','Methy_R','N6-ace_K','Palm_C','Phos_ST','Phos_Y','Pyro_Q','SUMO_K','Ubi_K','glyco_N','glyco_ST'. The following example code will run the saliency analysis on Phosphorylation on site 203 of protein P04150, and return a figure of surronding saliency scores.
python predict_saliency.py \
--inter \
--pretrain_name saved_model/MIND_fifteenfold \
--data_path sample/P04150.fa \
--res_path temp \
--site 203 \
--ptm_type Phos_STpython PTMSNP.py \
--pretrain_name saved_model/MIND_fifteenfold \
--data_path [path to fasta file] \
--res_path [path to store result] \
--snp [snp e.g. R_1022_C] \
--n_fold 15Please cite the following article for usage.
MIND-S is a deep-learning prediction model for elucidating protein post-translational modifications in human diseases.