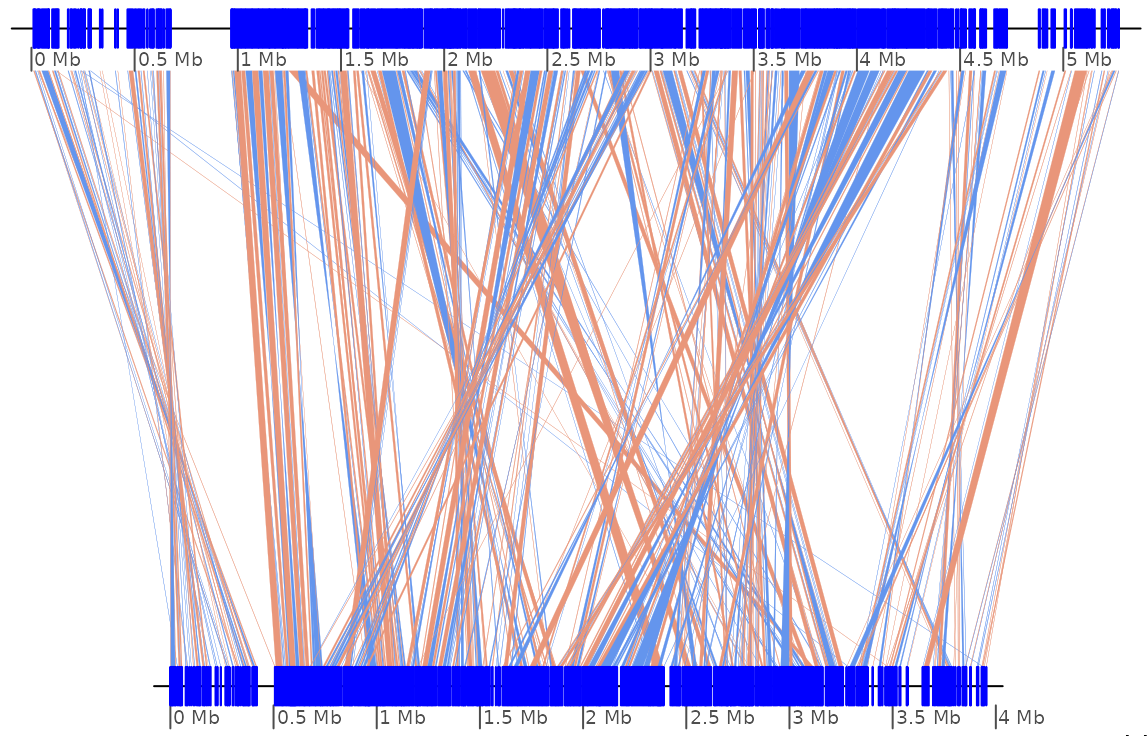
GenomicBreaks is a R package using Bioconductor
libraries to analyse pairwise alignments of whole genomes in which the gene
order has been scrambled by evolution, like in the picture below that represents
the comparison of homologous chromosomes in two distantly related molds,
N. crassa (chrIII) and P. comata (chr7).
This package is especially designed to parse and process the alignment files produced by the our pairwise genome alignment pipeline, but should be capable to import output of other pipelines as well.
The following should work:
Rscript -e 'remotes::install_github("oist/GenomicBreaks", repos=BiocManager::repositories())'
Add dependencies=TRUE if you would like to install the packages needed to build the vignettes.
On a Debian/Ubuntu system, try this:
sudo apt install r-base
sudo apt install pandoc qpdf texlive # For vignette builds and package checks
sudo apt install libxml2-dev libcurl4-openssl-dev libssl-dev libfftw3-dev libtiff-dev libgsl-dev
sudp atp install libfontconfig1-dev libharfbuzz-dev libfribidi-dev # For pkgdown
sudo apt install git bash-completion
sudo apt install libgl1 libnss3 libasound2 libxdamage1
wget https://download1.rstudio.org/desktop/bionic/amd64/rstudio-2021.09.0-351-amd64.deb
sudo apt --fix-broken -y install ./rstudio-2021.09.0-351-amd64.deb
Rscript -e 'install.packages("BiocManager")'
Rscript -e 'install.packages("tidyverse")'
Rscript -e 'install.packages("devtools")'
Rscript -e 'install.packages("remotes")'
Rscript -e 'remotes::install_github("oist/GenomicBreaks", repos=BiocManager::repositories(), dependencies=TRUE)'
A pairwise alignment of two genomes is loaded in GBreaks objects wrapping
the GRanges class. Here is an example:
GBreaks object with 11 ranges and 2 metadata columns:
seqnames ranges strand | query score
<Rle> <IRanges> <Rle> | <GRanges> <integer>
[1] chr1 11256821-11271214 - | Chr1:7699877-7713142 14394
[2] chr1 11271261-11272159 - | Chr1:7975442-7976321 899
[3] chr1 11272246-11274272 + | Chr1:7686802-7688942 2027
[4] chr1 11275227-11276200 - | Chr1:7491169-7492136 974
[5] chr1 11276902-11281111 - | Chr1:7850371-7855204 4210
[6] chr1 11281154-11281731 + | PAR:10891068-10891635 578
[7] chr1 11281946-11288799 + | Chr2:9359434-9367027 6854
[8] chr1 11288839-11299743 - | Chr1:10912857-10921537 10905
[9] chr1 11300902-11301564 - | Chr1:9597979-9599493 663
-------
seqinfo: 19 sequences from OKI2018_I69 genome
See “Get started” on https://oist.github.io/GenomicBreaks for further details.