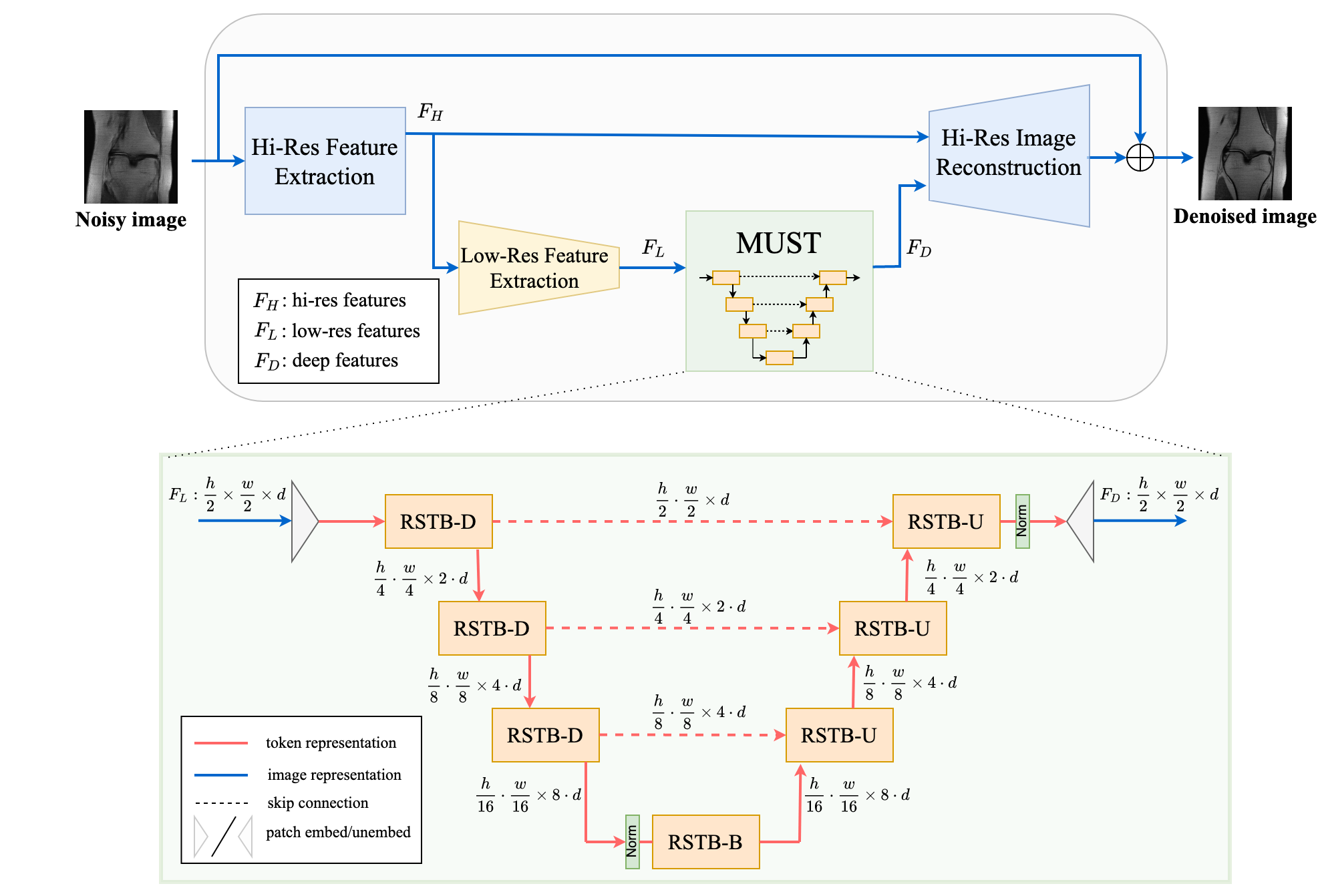
This is the PyTorch implementation of the NeurIPS 2022 paper HUMUS-Net, a Transformer-convolutional Hybrid Unrolled Multi-Scale Network architecture for accelerated MRI reconstruction.
HUMUS-Net: Hybrid unrolled multi-scale network architecture for accelerated MRI reconstruction,
Zalan Fabian, Berk Tınaz, Mahdi Soltanolkotabi
NeurIPS 2022
Reproducible results on the fastMRI multi-coil knee test dataset with x8 acceleration:
| Method | SSIM | NMSE | PSNR |
|---|---|---|---|
| HUMUS-Net (ours) | 0.8945 | 0.0081 | 37.3 |
| E2E-VarNet | 0.8920 | 0.0085 | 37.1 |
| XPDNet | 0.8893 | 0.0083 | 37.2 |
| Sigma-Net | 0.8877 | 0.0091 | 36.7 |
| i-RIM | 0.8875 | 0.0091 | 36.7 |
Pre-trained HUMUS-Net models for the fastMRI Public Leaderboard submissions can be found below.
This repository contains code to train and evaluate HUMUS-Net model on the fastMRI knee, Stanford 2D FSE and Stanford Fullysampled 3D FSE Knees datasets.
CUDA-enabled GPU is necessary to run the code. We tested this code using:
- Ubuntu 18.04
- CUDA 11.4
- Python 3.8.5
- PyTorch 1.10.1
First, install PyTorch 1.10.1 with CUDA support following the instructions here. Then, to install the necessary packages run
git clone https://github.com/z-fabian/HUMUS-Net
cd HUMUS-Net
pip3 install wheel
pip3 install -r requirements.txt
pip3 install pytorch-lightning==1.3.3FastMRI is an open dataset, however you need to apply for access at https://fastmri.med.nyu.edu/. To run the experiments from our paper, you need the download the fastMRI knee dataset with the following files:
- knee_singlecoil_train.tar.gz
- knee_singlecoil_val.tar.gz
- knee_multicoil_train.tar.gz
- knee_multicoil_val.tar.gz
After downloading these files, extract them into the same directory. Make sure that the directory contains exactly the following folders:
- singlecoil_train
- singlecoil_val
- multicoil_train
- multicoil_val
Please follow these instructions to batch-download the Stanford datasets. Alternatively, they can be downloaded from http://mridata.org volume-by-volume at the following links:
After downloading the .h5 files the dataset has to be converted to a format compatible with fastMRI modules. To create the datasets used in the paper please follow the instructions here.
To train HUMUS-Net on the fastMRI knee dataset, run the following in the terminal:
python3 humus_examples/train_humus_fastmri.py \
--config_file PATH_TO_CONFIG \
--data_path DATA_ROOT \
--default_root_dir LOG_DIR \
--gpus NUM_GPUSPATH_TO_CONFIG: path do the.yamlconfig file containing the experimental setup and training hyperparameters. Config files can be found in thehumus_examples/experimentsfolder. Alternatively, you can create your own config file, or directly pass all arguments in the command above.DATA_ROOT: root directory containing fastMRI data (with folders such asmulticoil_trainandmulticoil_val)LOG_DIR: directory to save the log files and model checkpoints. Tensorboard is used as default logger.NUM_GPUS: number of GPUs used in DDP training assuming single-node multi-GPU training.
Similarly, to train on either of the Stanford datasets, run
python3 humus_examples/train_humus_stanford.py \
--config_file PATH_TO_CONFIG \
--data_path DATA_ROOT \
--default_root_dir LOG_DIR \
--train_val_seed SEED \
--gpus NUM_GPUSIn this case DATA_ROOT should point directly to the folder containing the converted .h5 files. SEED is used to generate the training-validation split (0, 1, 2 in our experiments).
Note: Each GPU is assigned whole volumes of MRI data for validation. Therefore the number of GPUs used for training/evaluation cannot be larger than the number of MRI volumes in the validation dataset. We recommend using 4 or less GPUs when training on the Stanford 3D FSE dataset.
Here you can find checkpoint files for the models submitted to the fastMRI Public Leaderboard. See the next section how to load/evaluate models from the checkpoint files.
| Dataset | Model | Trained on | Acceleration | Checkpoint size | Link |
|---|---|---|---|---|---|
| fastMRI Knee | default | train | x8 | 1.4G | Download |
| fastMRI Knee | default | train+val | x8 | 1.4G | Download |
| fastMRI Knee | large | train | x8 | 2.8G | Download |
| fastMRI Knee | large | train+val | x8 | 2.8G | Download |
| Stanford 2D FSE | default | seed 0 split | x8 | 1.4G | Download |
| Stanford 3D FSE Knees | default | seed 0 split | x8 | 1.4G | Download |
To evaluate a model trained on fastMRI knee data on the validation dataset, run
python3 humus_examples/eval_humus_fastmri.py \
--checkpoint_file CHECKPOINT \
--data_path DATA_DIRCHECKPOINT: path to the model checkpoint.ckptfile
Note: by default, the model will be evaluated on 8x acceleration.
To evaluate on one of the Stanford datasets run
python3 humus_examples/eval_humus_stanford.py \
--checkpoint_file CHECKPOINT \
--data_path DATA_DIR \
--gpus NUM_GPUS \
--train_val_split TV_SPLIT \
--train_val_seed TV_SEEDTV_SPLIT: portion of dataset to be used as training data, rest is used for validation. For example if set to0.8(default), then 20% of data will be used for evaluation now.TV_SEED: seed used to generate the train-val split.
To experiment with different network settings see all available training options by running
python3 humus_examples/train_humus_fastmri.py --helpAlternatively, the .yaml files in humus_examples/experiments can be customized and used as config files as described before.
HUMUS-Net is MIT licensed, as seen in the LICENSE file.
If you find our paper useful, please cite
@article{fabian2022humus,
title={{HUMUS-Net}: Hybrid Unrolled Multi-scale Network Architecture for Accelerated {MRI} Reconstruction},
author={Fabian, Zalan and Tinaz, Berk and Soltanolkotabi, Mahdi},
journal={Advances in Neural Information Processing Systems},
volume={35},
pages={25306--25319},
year={2022}
}- fastMRI repository
- fastMRI: Zbontar et al., fastMRI: An Open Dataset and Benchmarks for Accelerated MRI, https://arxiv.org/abs/1811.08839
- Stanford 2D FSE: Joseph Y. Cheng, https://github.com/MRSRL/mridata-recon/
- Stanford Fullysampled 3D FSE Knees: Epperson K, Sawyer AM, Lustig M, Alley M, Uecker M., Creation Of Fully Sampled MR Data Repository For Compressed Sensing Of The Knee. In: Proceedings of the 22nd Annual Meeting for Section for Magnetic Resonance Technologists, 2013