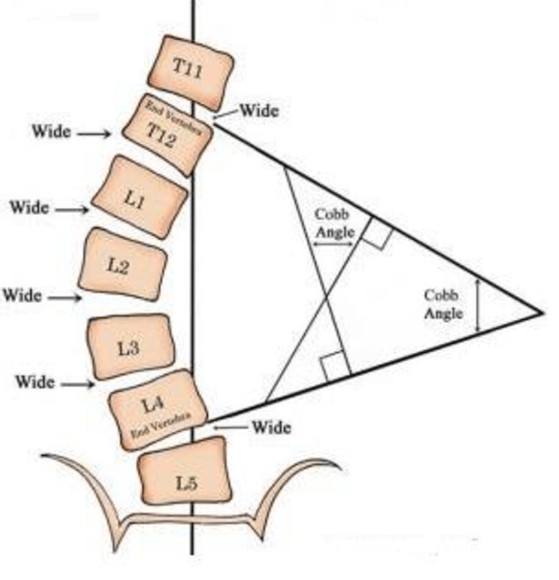
Cobb angle can be measured from spinal X-ray images.
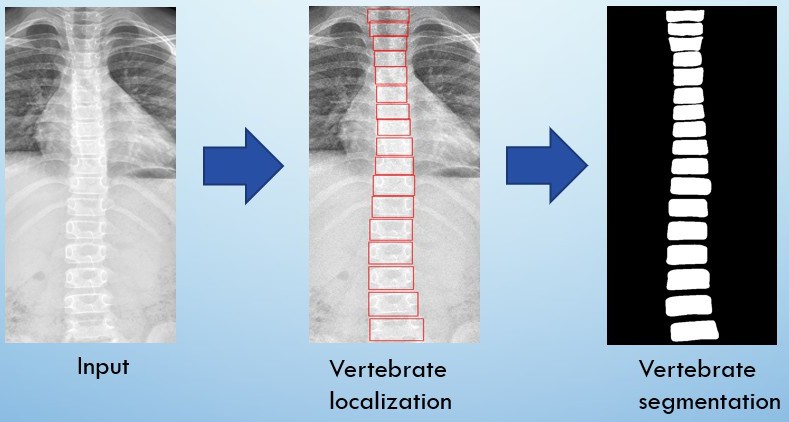
Automatic locate and segment the vertebra from an anterior-posterior (AP) view spinal X-ray images (grey level).
Explore the docs »
View Demo
·
Report Bug
·
Request Feature
Table of Contents
About The Project
Built With
Getting Started
To get a local copy up and running follow these simple steps.
Prerequisites
- Python
numpy
torch>=1.4
torchvision
matplotlib
tensorflow
tensorboard
terminaltables
pillow
tqdmInstallation
- Clone the repo
git clone https://github.com/z0978916348/Localization_and_Segmentation.git- Install python packages
pip install requirements.txt- Prepare VertebraSegmentation data
└ train
└ 1.png
└ 2.png
└ ...
└ test
└ 5.png
└ 6.png
└ ...
└ valid
└ 8.png
└ 9.png
└ ...- Prepare PyTorch_YOLOv3 data
Classes
Add class names to data/custom/classes.names. This file should have one row per class name.
Image Folder
Move the images of your dataset to data/custom/images/.
"train and valid respectively"
Annotation Folder
Move your annotations to data/custom/labels/. The dataloader expects that the annotation file corresponding to the image data/custom/images/train.jpg has the path data/custom/labels/train.txt. Each row in the annotation file should define one bounding box, using the syntax label_idx x_center y_center width height. The coordinates should be scaled [0, 1], and the label_idx should be zero-indexed and correspond to the row number of the class name in data/custom/classes.names.
"train and valid respectively"
Define Train and Validation Sets
In data/custom/train.txt and data/custom/valid.txt, add paths to images that will be used as train and validation data respectively.
After training, you can follow app.py folder to put your dataset into correspond folder. It it easy to know, I will not step by step for description
- Excute
python app.pyUsage
Use deep learning model to detect and segment spine precisely. We can use it to find Cobb Angle
Flow Chart
Contributing
Contributions are what make the open source community such an amazing place to be learn, inspire, and create. Any contributions you make are greatly appreciated.
- Fork the Project
- Create your Feature Branch (
git checkout -b feature/AmazingFeature) - Commit your Changes (
git commit -m 'Add some AmazingFeature') - Push to the Branch (
git push origin feature/AmazingFeature) - Open a Pull Request
License
Distributed under the MIT License. See LICENSE for more information.
Contact
Project Link: https://github.com/z0978916348/Localization_and_Segmentation
References
- Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
- Z. Zhang, Q. Liu, and Y. Wang, “Road extraction by deep residual u-net,” IEEE Geoscience and Remote Sensing Letters, 2018, http://arxiv.org/abs/1711.10684.
- Horng, M. H., Kuok, C. P., Fu, M. J., Lin, C. J., & Sun, Y. N. (2019). Cobb Angle Measurement of Spine from X-Ray Images Using Convolutional Neural Network. Computational and mathematical methods in medicine, 2019.
- Al Arif, S. M. R., Knapp, K., & Slabaugh, G. (2017). Region-aware deep localization framework for cervical vertebrae in X-ray images. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support (pp. 74-82). Springer, Cham.