Moments is the site frequency spectrum (SFS) analysis method superficially similar to dadi but operating on different math (ordinary differential equations rather than diffusion approximation). It is considerably faster than dadi (although somewhat less accurate), handles up to five populations simultaneously, and plots cartoons of inferred demographic scenarios.
The problem with moments and dadi is, they can evaluate the fit of a pre-specified demographic model but are not designed to search for the general model structure that best fits the data (i.e., was there population split or not, is there any migration and if yes, were there additional growth periods before or after spit, etc). Our solution to this problem is pretty simple: to fit all the two-population models we can possibly think of to our experimental 2dSFS and use Akaike Information Criterion to select the best one.
See GADMA for the alternative solution to this problem. Compared to GADMA, we are far less elegant but somewhat more flexible (we can incorporate essentially any model, including models involving background selection and heterogeneous introgression rates across the genome). Our approach also lets the user evaluate how much better the winning model is compared to certain "null" alternatives (for example, models with no population split or with constant population sizes), which provides statistical evidence for general aspects of the model structure. Our disadvantage (besides the fact that we only do two-pop models and GADMA also does three-pop models) is a huge number of model-fit runs that we have to perform. The good news is, all this can be done in parallel.
Version 2 (still very beta! use at your own risk) of this repository (multimodel_inference/py3_v2) actually uses GADMA genetic algorithm for find optimal parameters of a pre-specified model, which is supposed to make it much more robust. It also features a completely revamped set of models - they can be used without GADMA as "v.2 vanilla". The new models don't have versions with symmetrical migration (like in version 1), but include models with "background selection" (reduced Ne in a portion of the genome). Both version 1 and 2 have models with "islands of divergence" (reduced migration in a portion of the genome).
Rippe, John P., Groves Dixon, Zachary L. Fuller, Yi Liao, and Mikhail Matz. 2021. “Environmental Specialization and Cryptic Genetic Divergence in Two Massive Coral Species from the Florida Keys Reef Tract.” Molecular Ecology, https://doi.org/10.1111/mec.15931.
First of all, install moments. The example below would clone it into the user's home directory and install it for a specific user.
cd
git clone --branch devel https://bitbucket.org/simongravel/moments.git
cd moments
python3 setup.py build_ext --inplace
cd
# add moments to $PYTHONPATH (consider adding this line to your .bashrc):
export PYTHONPATH=$PYTHONPATH:$HOME/momentsThen, (if you want to try v.2 with GADMA) install GADMA:
# if you are root user:
sudo python3 -m pip install numpy
sudo python3 -m pip install gadma
# if not:
python3 -m pip install --user numpy
python3 -m pip install --user gadma
export PYTHONPATH=$PYTHONPATH:$HOME/.local/binThen, clone this repository and copy all the scripts and accessory filed for the version you'd like to work with into a newly created subdir work:
cd
git clone https://github.com/z0on/AFS-analysis-with-moments.git
cd AFS-analysis-with-moments
mkdir work
# to use version 1 models (RECOMMENDED):
cp multimodel_inference/py3_v1/* work/
# to use v.2 models without GADMA engine (BETA! If results don't make sense please do tell me)
# cp multimodel_inference/py3_v2/vanilla/* work/
# to use full on v.2 with GADMA (BETA! If results don't make sense please do tell me)
# cp multimodel_inference/py3_v2/GA/* work/
# to use version 1 models for python2
# cp multimodel_inference/py2_v1/* work/NOTE: all code examples here assume the repository is cloned in the home directory,
~/. If you cloned it elsewhere, make sure to replace~/AFS-analysis-with-momentsin all examples with the actual path.
NOTE: R scripts were tested with R versions 3.5.1 and 3.6.3. Not sure about R version 4.
- The first step is model selection, where we run all possible models on 10 bootstrapped SFS. We run each model on each bootstrap six times (six random restarts), to make sure the model converges to its best likelihood at least once. All these commands are written by the
RscriptmodSel_write.R. Then we use theRscriptmodSel_summary.Rto select the best-fitted instance (out of 6) for each model for each bootstrap, and compare the AIC scores for all models. The best model is the one with the lowest median AIC score among bootstrap replicates. - The second step is running the winning model on 100 bootstrapped SFS, to evaluate parameter uncertainties. The commands for this stage are actually written by the
modSel_summary.Rscript. Once again, we are doing 6 random restarts for each bootstrap replicate. The parameter meanings and uncertainties are deciphered by the thirdRscript that we have,bestBoot_summary.R. All threeRscripts are designed for command-line usage.
The models are designed to test the following basic aspects of population configuration:
- are these really two demographically distinct populations, or we simply sampled the same population twice? (i.e., does the model fits significantly better if it actually has a split between populations, as opposed to just some population size changes)
- were there changes in population size(s) through time? (models can include up to three "epochs" where population size could change)
- if there is a split, is there still migration? (during some or all of the epochs)
- (version 1 only) if there is migration, is it symmetric or asymmetric?
- do some parts of the genome ("islands of differentiation") introgress at a lower rate than the rest? This is one way to model non-neutral processes such as spatially varying selection.
- (version 2 only) do some parts of the genome show lower population size than the rest? This is to model background selection.
So the models differ by:
- split / no split (
nsin the model name) - no migration ever (
nm), or some migration (all other models) - number of epochs (1-3) (
s1,s2ors3) - migration at some or all of the epochs (there are models with secondary contact,
sc, or ancestral migration,am) - models with ancestral population size change before split (
12with one epoch post-split,123with two epochs post-split,103is the secondary contact version of123- no migration in the middle epoch) - presence of "islands of differentiation" (
i) - (version 2 only) presence of islands of "background selection" (
S).
NOTE for version 1: the model names are not fully standardized to the above convention, please see
work/moments_multimodels.xlsxfor their structure.
NOTE for version 2:
IMmodels are currently not included in the main collection of models since they take substanitally longer to fit (some can take 4-5 hours). All v.2 IM models are ofmnekind, which means that migration scales dynamically with the size of the source population. If you want to include them, copy the extended model lists over the standard ones:
cp ~/AFS-analysis-with-moments/work/allmodels_IMextra_unfolded ~/AFS-analysis-with-moments/work/allmodels_unfolded
cp ~/AFS-analysis-with-moments/work/allmodels_IMextra_folded ~/AFS-analysis-with-moments/work/allmodels_foldedSee the spreadsheet work/moments_multimodels.xlsx (v.1) or work/multimodels_v2.xlsx (v.2) for summaries of model structure.
Let's assume we have ten bootstrapped 2dSFS formatted for moments or dadi (See Appendix for instructions how to obtain bootstrapped 2dSFS from ANGSD). Such file is nothing more than a line of numbers with a header line giving the dimensions of the spectrum ( 2 x N + 1 for each of the two populations, where N is the number of sampled diploids).
Bootstrapped SFS files should be named like p12_1.sfs, p12_2.sfs, etc. where p12 is the name of population contrast.
To create a list of AFS models to run, do this:
cd [where your boostrapped SFS files are]
Rscript ~/AFS-analysis-with-moments/work/modSel_write.R contrast=p12 args="p1 p2 16 16 0.02 0.005"where
contrast: the name of population comparison. It should match the leading part of the bootstapped SFS names (in example here,p12)args: list of parameters for AFS models, in the following order: name of pop1, name of pop2, projection for pop1, projection for pop2, mutation rate (per genotyped portion of the genome per generation), generation time in thousands of years.
Population names can be anything. For ANGSD-derived SFS, projections should be 1.6N for each population (rounded to integer); in the case shown here, each population was represented by 10 individuals.
Additional arguments to modSel_write.R (defaults):
nreps(6 for v.1, 3 for v.2) : number of random restarts for each model for each bootstrap rep.nboots(10) : number of bootstrap replicates to use. 10 seems to be optimal at this stage.path2models(~/AFS-analysis-with-moments/multimodel_inference/) : path to where the package livesfolded(FALSE) : whether to fold the SFS for analysis
Note: if you want to analyze folded SFS, generate unfolded ones and specify
folded=TRUEhere and at the next stage.
Run all commands in [contrast].modsel.runs file. This is the most computaitonally intensive thing I have ever done - in version 1, there are 6 x 108 x 10 model runs, requiring 1 hour each. Best run these on an HPC cluster, in parallel! All the screen output is going to be collected in a file, p12.modsel in this case.
Note: some model runs may not finish in 1 hour; just kill them. These are hopeless runs where the parameter search algorithm is stuck, they will have horrible fit even if they eventually finish.
Then, to summarize results and write the list of commands for the next step (bootstrapping the winnign model):
Rscript ~/AFS-analysis-with-moments/work/modSel_summary.R modselResult=p12.modsel args="p1 p2 16 16 0.02 0.005"where
modselResult: the name of the resulting file from model selection, typically[contrast].modsel.args: same argument as formodSel_write.R
Additional arguments to modSel_summary.R that will influence the next stage, bootstrapping the winning model (defaults):
nreps(6) : number of random restarts for each model for each bootstrap rep.nboots(100) : number of bootstrap replicates.folded(FALSE) : whether analysis is using folded SFS.
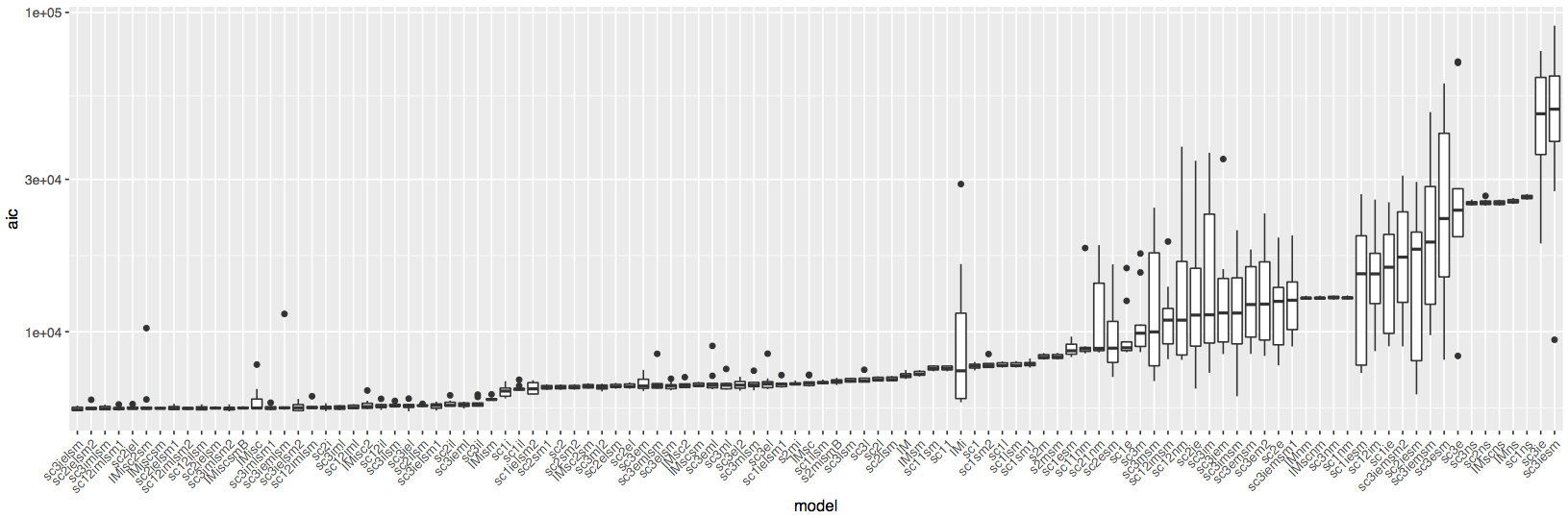
Two plots will be generated. The first one is the boxplot of best AIC scores for each model for all bootstrap replicates:
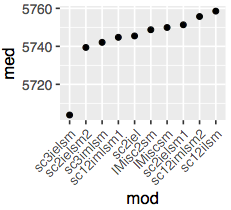
And the second one is the plot of just the AIC medians for the top 10 models:
The script also outputs the text file named [contrast].[modelname], where [modelname] is the name of the winning model. This file contains the fitted parameter values for the winning model, which will be used at the next stage as "guiding values" for random restarts.
Lastly, the script outputs a file [contrast].winboots.runs that contains all the commands to run the next stage.
Assuming we have 100 boostrapped SFS (See Appendix for instructions how to obtain bootstrapped 2dSFS from ANGSD), we are now going to run just the winning model on all of them. The commands file to do that has been already created by running modSel_summary.R, in the current example it is the file p12.winboots.runs. Now all we need to do is to run all these commands, much preferably in parallel. As before, let them run for one hour, kill all that did not finish.
The text output will be collected in the file p12.winboots. To summarize it all we need to do is
Rscript ~/AFS-analysis-with-moments/work/bestBoot_summary.R bootRes=p12.winbootsAdditonal options to bestBoot_summary.R are:
topq: top quantile cutoff. Only boostrap runs in this top quantile will be summarized. Default 0.5path2models: path to the subdirmultimodel_inference. Default~/AFS-analysis-with-moments/multimodel_inference/.folded(FALSE) : whether the analysis must fold the SFS.
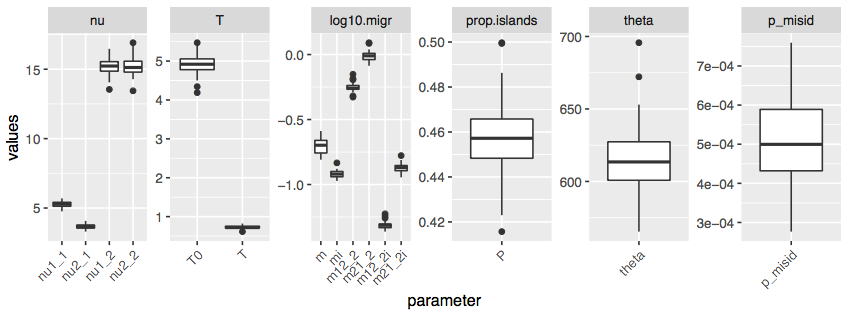
This will generate the boxplots for parameter estimates:
Note: names of migration rate parameters are not entirely systematized across models (I should spend some time cleaning those). In general the higher number (i.e.
_3inm12_3) indicates later epoch, but they may not exactly follow the number of epochs actually present in the model.
The script also saves an RData bundle containing the summary dataframe (medians, 25% quantile, 75% quantile for all parameters) and the big dataframe containing all summarized bootstrap data.
Last but not least, the script identifies the model run that is the closest to the median across bootstrap replicates and saves two plots for it:
[input filename]_representativeModel.png: model structure[input filename]_representativeModel.pdf: comparison of real and model-derived SFS.
Here we obtain 100 series of 5 block-bootstrap replicates, which we then average. This averaging procedure is called "bagging" and is meant to mitigate the noise that ANGSD-derived SFS often show in the area of high-frequency (implying low count) variants. The resulting 100 "bagged" datasets are going to be our bootstrap replicates.
Let's assume we have two populations, p1 and p2, each with 10 sequenced individuals, and we have two text files, p1.bams and p2.bams, listing *.bam files for each population. We will make site frequency spectra from them while applying quality and genotyping rate filters, but NOT the filters that rely on commonality of the variant (such as minMaf or snp-pval). This is because we want to keep all the well-genotyped sites, variable (including singletons) and invariable.
Note: One of the filters we need to use is minInd - minimum number of individuals the site should be genotyped at. We want to keep the genotyping rate (the proprotion of all individuals in which the site is genotyped) the same in the two populations, so we will have to do a bit of silly bash arithmetics in the beginning of the following code chunk.
export GenRate=0.75 # desired genotyping rate
export N1=`wc -l p1.bams | cut -f 1 -d " "`
export N2=`wc -l p2.bams | cut -f 1 -d " "`
export MI1=`echo "($N1*$GenRate+0.5)/1" | bc`
export MI2=`echo "($N2*$GenRate+0.5)/1" | bc`
FILTERS='-uniqueOnly 1 -skipTriallelic 1 -minMapQ 30 -minQ 30 -maxHetFreq 0.5 -hetbias_pval 1e-3'
# add `-sb_pval 1e-3` (strand bias) to FILTERS if you have 2bRAD, GBS, or WGS data. Other types of RAD only sequence one strand so -sb_pval filter would remove everything.
GENOME_REF=mygenome.fasta # reference to which the reads were mapped
TODO="-doHWE 1 -doSaf 1 -doMajorMinor 1 -doMaf 1 -doPost 1 -dosnpstat 1 -anc $GENOME_REF -ref $GENOME_REF"
angsd -b p1.bams -GL 1 -P 4 -minInd $MI1 $FILTERS $TODO -out p1.0
angsd -b p2.bams -GL 1 -P 4 -minInd $MI2 $FILTERS $TODO -out p2.0
# collecting and indexing filter-passing sites in each population
zcat p1.0.mafs.gz | cut -f 1,2 | tail -n +2 | sort >p1.sites
zcat p2.0.mafs.gz | cut -f 1,2 | tail -n +2 | sort >p2.sites
# collecting and indexing common sites:
comm -12 p1.sites p2.sites | sort -V >allSites
angsd sites index allSites
# listing "regions"
cat allSites | cut -f 1 | uniq >regions
# estimating site frequency likelihoods for each population
GENOME_REF=mygenome.fasta
angsd -rf regions -sites allSites -b p1.bams -GL 1 -doSaf 1 -anc $GENOME_REF -out p1
angsd -rf regions -sites allSites -b p2.bams -GL 1 -doSaf 1 -anc $GENOME_REF -out p2Note: don't worry about folding at this point. We will fold the spectra later, when running moments models, if needed.
Now we generate the bootstrapped data (100 series of 5 bootstraps):
export GENOME_REF=mygenome.fasta # reference to which the reads were mapped
>b100
for B in `seq 1 100`; do
echo "sleep $B && realSFS p1.saf.idx p2.saf.idx -ref $GENOME_REF -anc $GENOME_REF -bootstrap 5 -P 1 -resample_chr 1 >p12_$B">>b100;
done
Execute all commands in b100 - this will take a while so better do it in parallel.
Finally, we do "bagging" (averaging of 5 bootstrap replicates within each of the 100 series):
# computing SFS dimensions
export N1=`wc -l p1.bams | cut -f 1 -d " "`
export N2=`wc -l p2.bams | cut -f 1 -d " "`
export NG1=`echo "($N1*2)+1" | bc`
export NG2=`echo "($N2*2)+1" | bc`
# averaging 5-bootstrap batches
for B in `seq 1 100`; do
echo"$NG1 $NG2" >p12_${B}.sfs;
cat p12_${B} | awk '{for (i=1;i<=NF;i++){a[i]+=$i;}} END {for (i=1;i<=NF;i++){printf "%.3f", a[i]/NR; printf "\t"};printf "\n"}' >> p12_${B}.sfs;
done
And voila, we have 100 "bagged" bootstrapped SFS spectra named p12_1.sfs, p12_2.sfs, ..., p12_100.sfs.
Link to original Moments paper
Moments manual (link may change with updates)