conda env create -f pocketgen.yaml
conda activate pocketgenconda create -n targetdiff python=3.8
conda activate targetdiff
conda install pytorch pytorch-cuda=11.6 -c pytorch -c nvidia
conda install pyg -c pyg
conda install rdkit openbabel tensorboard pyyaml easydict python-lmdb -c conda-forge
conda install -c conda-forge openmm pdbfixer flask
conda install -c conda-forge numpy swig boost-cpp sphinx sphinx_rtd_theme
pip install meeko==0.1.dev3 wandb scipy pdb2pqr vina==1.2.2
python -m pip install git+https://github.com/Valdes-Tresanco-MS/AutoDockTools_py3We use CrossDocked and Binding MOAD datasets to benchmark pocket generation.
We download and process the CrossDocked dataset as described by the authors of TargetDiff
Firstly download the crossdocked_v1.1_rmsd1.0.tar.gz and split_by_name.pt and put it under the ./data directory.
Use the following commands to extract pockets, create index_seq.pkl, and split the dataset.
python data_preparation/extract_pockets.py
python data_preparation/split_pl_dataset.py
We download and process the Binding MOAD dataset following the authors of DiffSBDD Download the dataset
wget http://www.bindingmoad.org/files/biou/every_part_a.zip
wget http://www.bindingmoad.org/files/biou/every_part_b.zip
wget http://www.bindingmoad.org/files/csv/every.csv
unzip every_part_a.zip
unzip every_part_b.zipProcess the raw data using
python -W ignore process_bindingmoad.py <bindingmoad_dir>Use the following commands to extract pockets, create index_seq.pkl, and split the dataset.
python data_preparation/extract_pockets_moad.py
python data_preparation/split_pl_dataset_moad.py
We also provide the processed datasets for training from scratch at zenodo
For each dataset, it requires the preprocessed .lmdb file and split file _split.pt
Benchmarking PocketGen and other approaches for pocket generation on two datasets. Reported are average and standard deviation values across three independent runs. The best results are bolded.
| Model | AAR (↑) CrossDocked | Designability (↑) CrossDocked | Vina (↓) CrossDocked | AAR (↑) Binding MOAD | Designability (↑) Binding MOAD | Vina (↓) Binding MOAD |
|---|---|---|---|---|---|---|
| Test set | - | 0.77 | -7.016 | - | 0.79 | -8.076 |
| DEPACT | 31.52±3.26% | 0.68±0.04 | -6.632±0.18 | 35.30±2.19% | 0.67±0.06 | -7.571±0.15 |
| dyMEAN | 38.71±2.16% | 0.71±0.03 | -6.855±0.06 | 41.22±1.40% | 0.70±0.03 | 0.71±0.04 |
| FAIR | 40.16±1.17% | 0.73±0.02 | -7.015±0.12 | 43.68±0.92% | 0.72±0.05 | -7.930±0.15 |
| RFDiffusion | 46.57±2.07% | 0.74±0.01 | -6.936±0.07 | 45.31±2.73% | 0.75±0.05 | -7.942±0.14 |
| RFDiffusionAA | 50.85±1.85% | 0.75±0.03 | -7.012±0.09 | 49.09±2.49% | 0.78±0.03 | -8.020±0.11 |
| PocketGen | 63.40±1.64% | 0.77±0.02 | -7.135±0.08 | 64.43±2.35% | 0.80±0.04 | -8.112±0.14 |
Train on CrossDocked:
python train_recycle.py --config ./config/train_model.yml
Train on Binding MOAD:
python train_recycle.py --config ./config/train_model_moad.yml
Pretrained checkpoint on the CrossDocked training dataset: checkpoint.pt
python generate_new.py
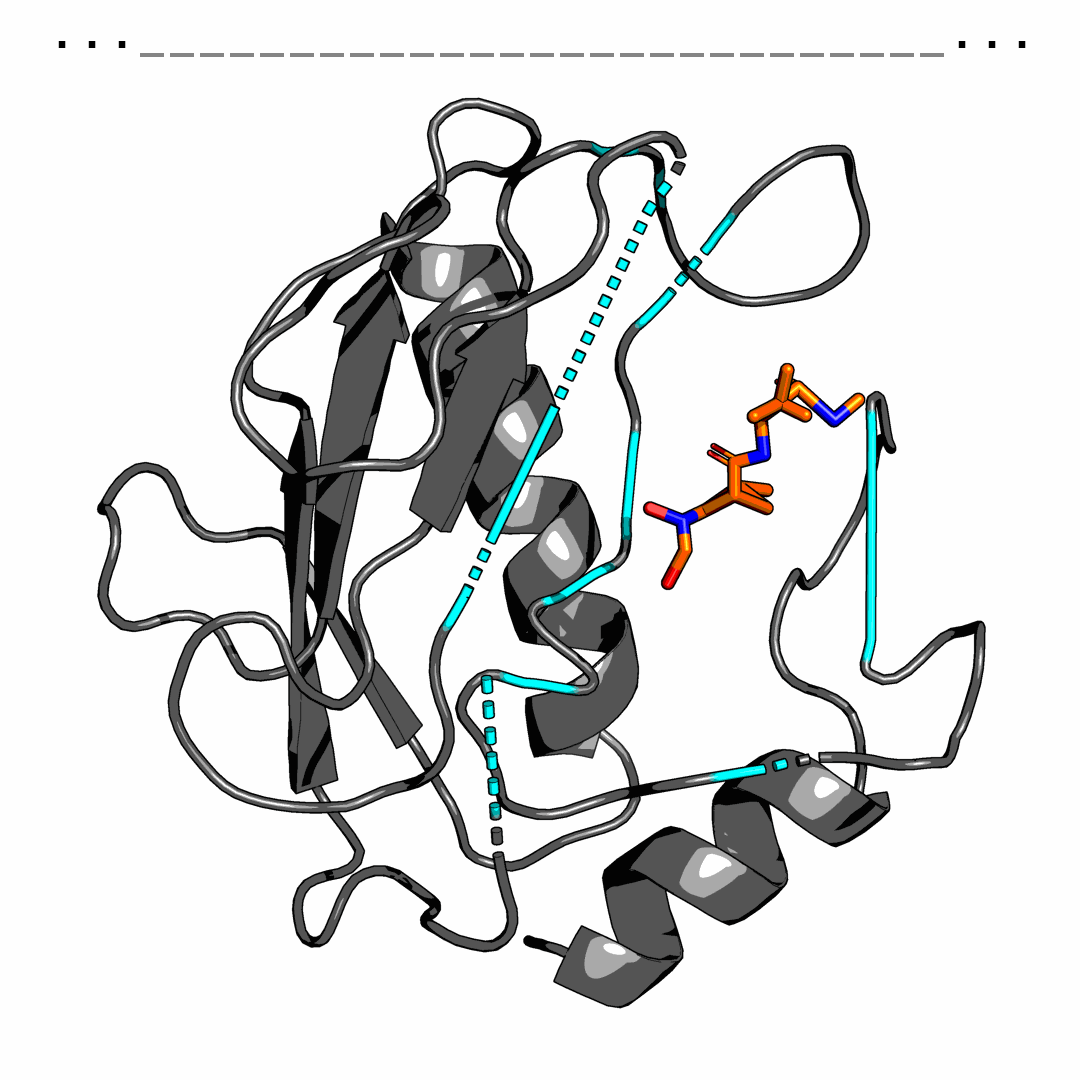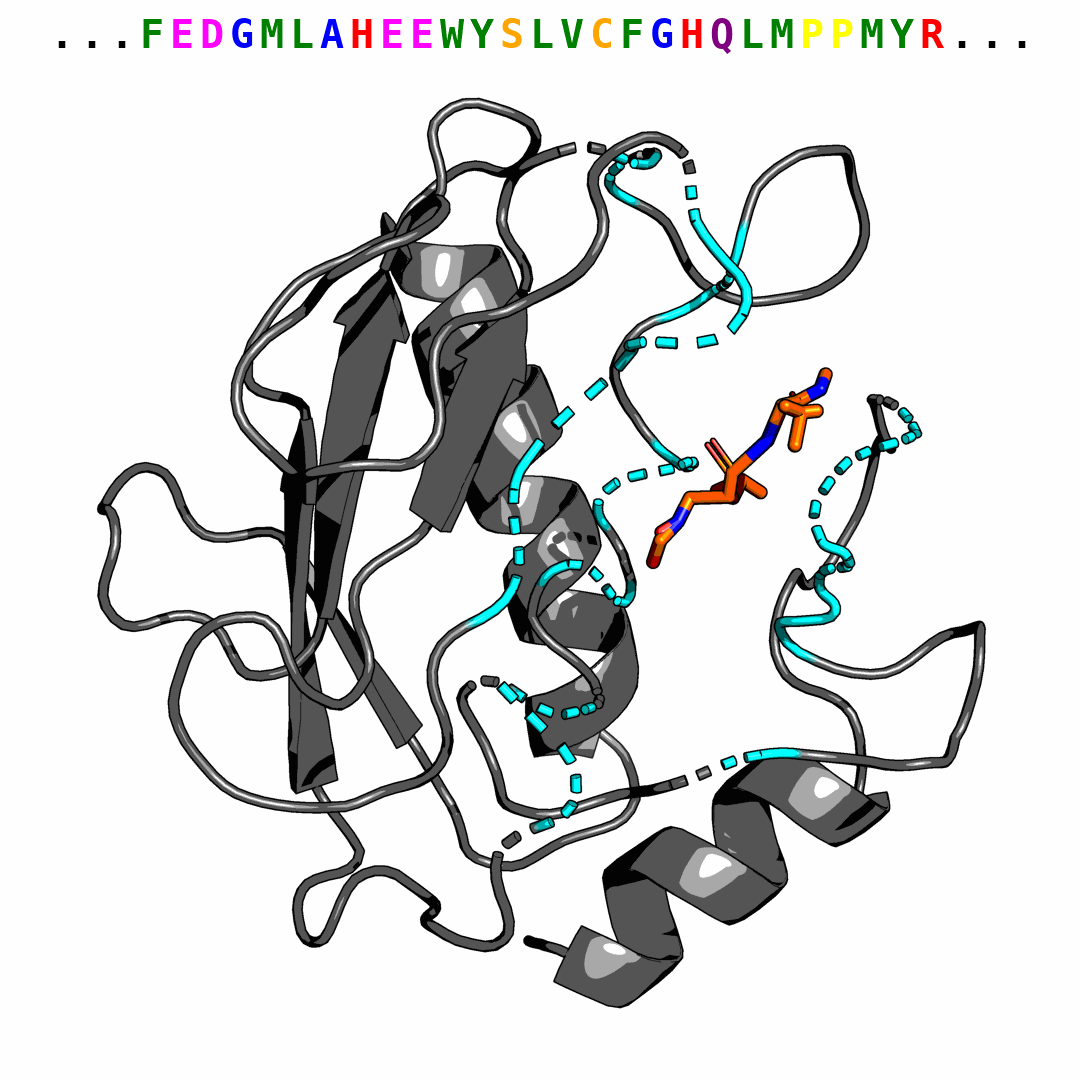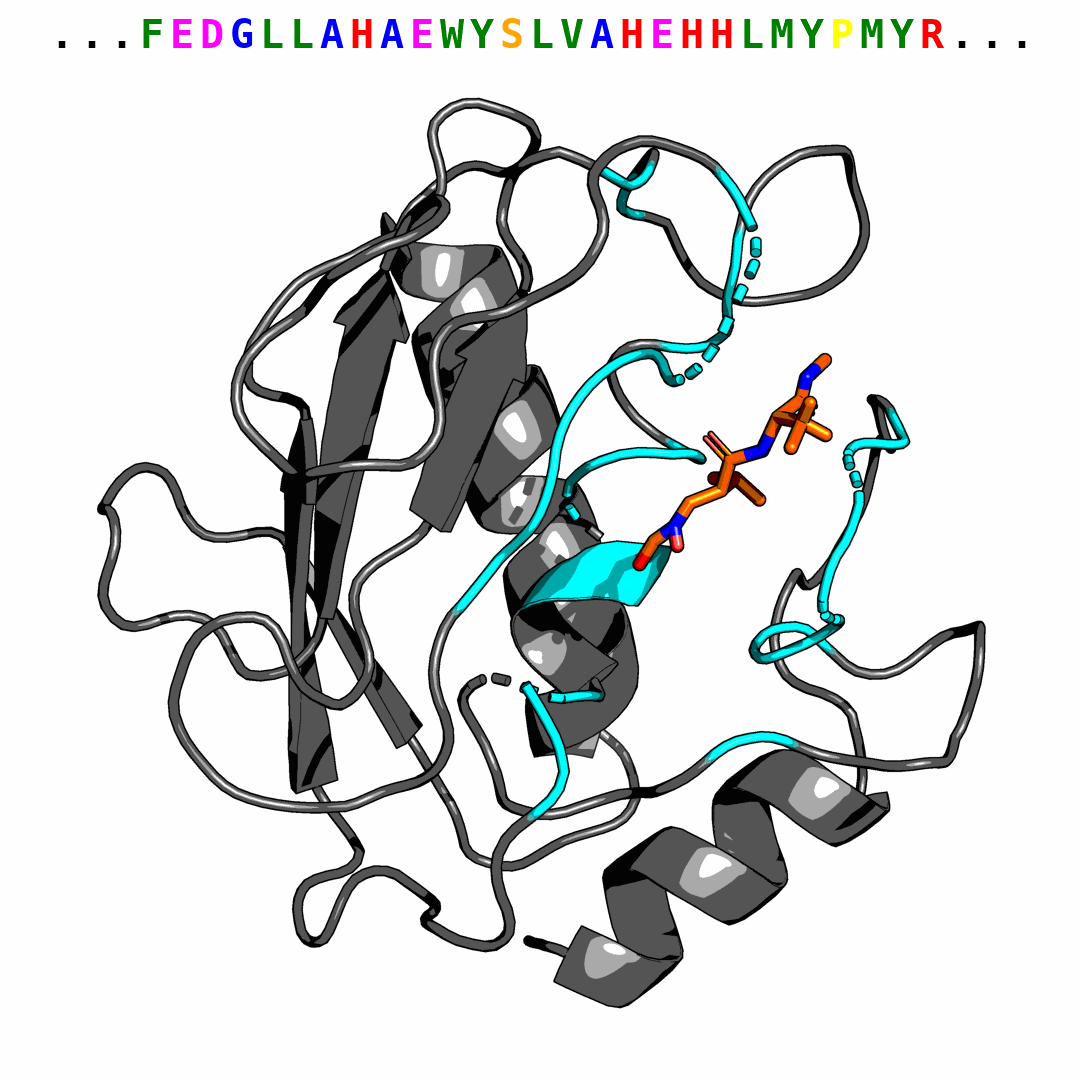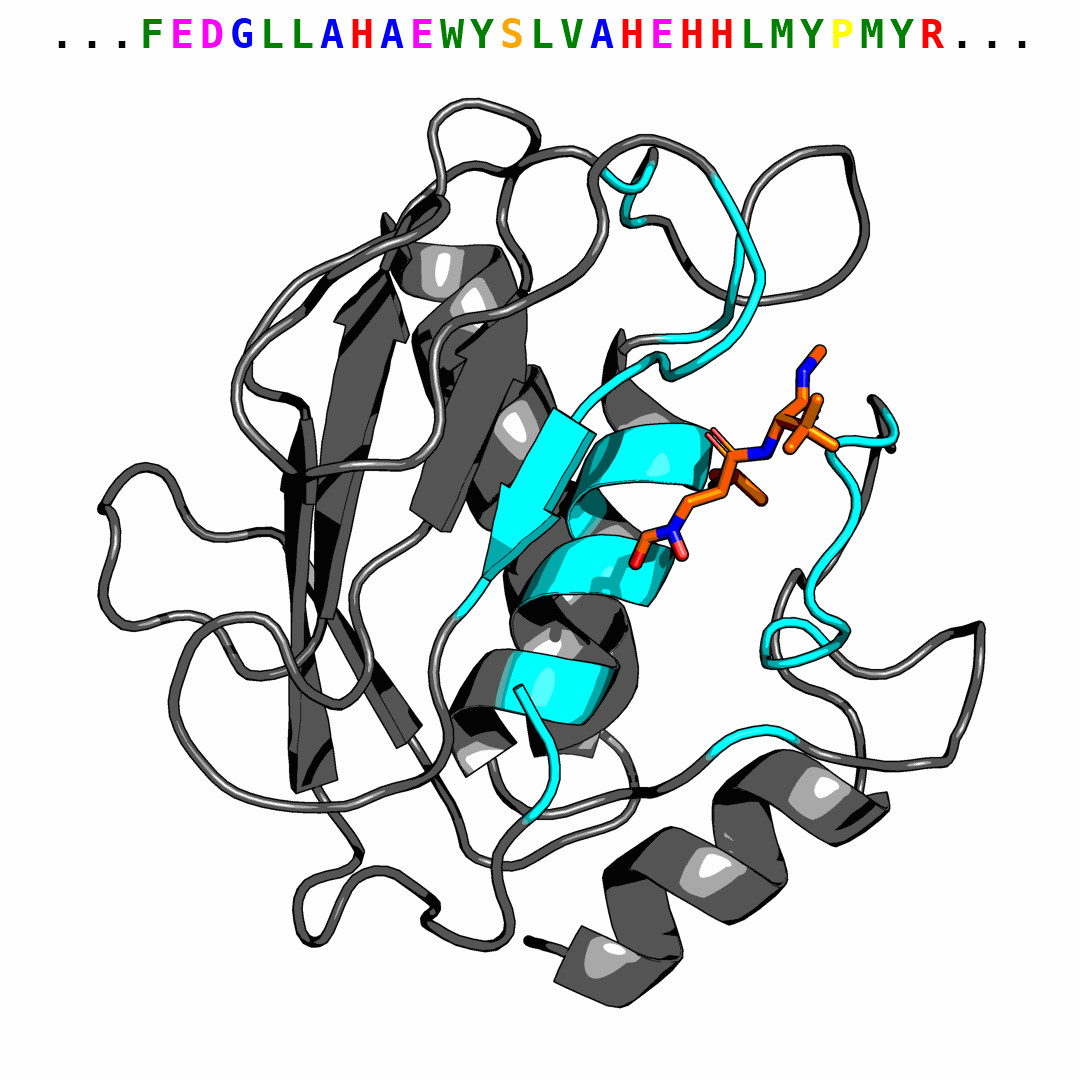
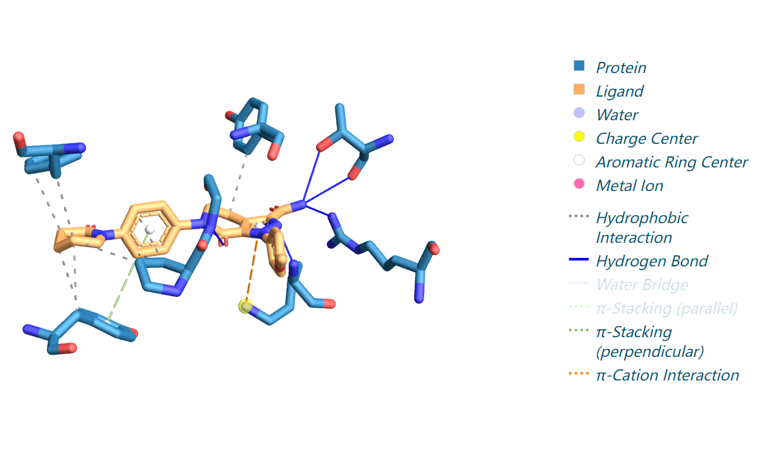
We provide one example of the generated pocket for pdbid-2p16 and visualize the interactions with plip
For generation, please create a tmp dir under the running fold.
The code to compute self-consistency-related scores, such as scRMSD, scTM, and pLDDT can be found at eval.
The code to run protein-ligand interaction analysis is interaction.
This project draws in part from TargetDiff and ByProt, supported by MIT License and Apache-2.0 License. Thanks for their great work and code!
Zaixi Zhnag (zz8680@princeton.edu)
Sincerely appreciate your suggestions on our work!
This project is licensed under the terms of the MIT license. See LICENSE for additional details.
@article{zhang2024efficient,
title={Efficient generation of protein pockets with PocketGen},
author={Zhang, Zaixi and Shen, Wan Xiang and Liu, Qi and Zitnik, Marinka},
journal={Nature Machine Intelligence},
pages={1--14},
year={2024},
publisher={Nature Publishing Group UK London}
}